Big Data and machine learning: new frontier in lung cancer care
Introduction
Lung cancer is one of the types of the cancer with very high incidence rate (1.61 million cases were recorded in 2008; 12.7% of the total cancer incidents) (1) and is one of the leading cancer killers across the world in both genders (1.38 million deaths were recorded in 2008; 18.2% of the total cancer lethality) (2). Common lung cancer causes include: smoking (3), exposure to second-hand smoke (4), exposure to radon gas (5,6), exposure to asbestos and other chemicals including air pollution (7-9), life style (lack of exercise, unhealthful diet, excess alcohol) (10), and family history of lung cancer. In the UK alone, it was estimated that more than 86% of lung cancers in women and around 91% in men are connected to the lifestyle, as well as environmental influences, showing that smoking is highly correlated with the cancer existence (11). Although smoking is considered to be one of the main risk factors, it was found that 10–15% of all patients with lung cancer have never smoked (11,12).
There are two main structures of lung cancer:
- non-small cell lung cancer (around 85% of all the lung cancers);
- small-cell lung cancer (around 15% of all the lung cancers).
Despite the advances in screening, early detection imaging and treatment improvement, the survival rate in patients with lung cancer prevails to be poor. The major reason for the devastating statistics of the lung cancer is the shortage of the approved tests able to detect small lung cancers, which are possible to be removed by surgery. Because of this, a number of preclinical and clinical research is done with the goal to optimize current approaches for both lung cancer types. The end goal is to optimize the outcomes by addressing the issues of prognostic and predictive factors which can affect the treatment outcome.
The importance of lung cancer analysis and treatment can be seen in many projects that are currently running or are finished; with the aim to understand the prediction of cancer development better and prescribe the adequate therapy. Some of the most influential projects are:
- REQUITE: Validating Predictive Models and Biomarkers of Radiotherapy Toxicity to Reduce Side-Effects and Improve Quality of Life in Cancer Survivor (13);
- LungCARD—Blood test for clinical therapy guidance of non-small cell lung cancer patients (14);
- RASTARGET—Targeting RAS oncogene addiction (15);
- PREDECT—New Models for Preclinical Evaluation of Drug Efficacy in Common Solid Tumours (16).
Concept of personalized medicine, where therapeutic decisions are created based on the genetic and histologic characteristics of the cancer, has been a significant improvement to a standard lung cancer diagnosis and treatment. Lung cancer molecular profiling has been nowadays improved by the advancements in scientific and technological cancer genome research. Cancer gene analysis of mutations has moved from the analysis of single gene to the analysis of next-generation global genome sequencing, meaning whole cancer genome sequencing. The growing magnitude of information about genomics is forecasted to improve our current knowledge of lung cancer significantly and would lead to the use of personalized lung cancer therapy as a standard therapy. Based on the current genetic and genomic database of lung cancer, researchers have been able to understand better the differences with respect to the lung cancer genetics, different human races crosswise. These results could potentially lead to improved treatment algorithm and therapeutic choices.
Lung cancer prevention and treatment—the role of ‘omics’ data
Researchers have found strong links between lung cancer mortality and gender, lifestyle factors, environmental factors and socioeconomic factors (17). In this age, where a number of targeted therapy and daily tests for predictive or prognostic molecular markers for many diseases are available, the availability of ‘omics’ data has high potential to be used for the lung cancer.
Biomarkers obtained from blood are a main evaluation target, as the blood is obtained daily in primary care clinics, with the minimal risk. Biggest efforts are focused on identification of genomics (genes) and proteomics (proteins) discharged by the tumor into blood. However, there are many challenges. The main challenge is the large molecular heterogeneity that can be found even for histologically similar tumors that may maintain disruption of different components of similar pathways, alterations to different cellular pathways, as well as unique disruption mechanisms to genes or pathways.
Besides the tumor variability, another big challenge is the fact that proteins and genes can provide only a small “image” of a tumor’s existence in the human body. The analysis of data requires a multi-omics systems-based approach that addresses several omics data types from each cancer specimen individually, followed then by integration of the mentioned dimensions in order to identify the primary genes and pathways that drive the associated phenotype (18) as well as tumor biomarkers that will enable prevention and early treatment.
Currently, two large international research (International Cancer Genome Consortium - ICGC and The Cancer Genome Atlas - TCGA) efforts are compiling ‘omics’ data for several cancer types. Their goal is to compile openly available ‘omics’ in order to improve our understanding of the molecular mechanisms driving cancer (19,20). Beside these two, there are also the NIH Roadmap Epigenomics Mapping Consortium (EMC), public resource that contains epigenomics maps for stem cells and normal tissues (21), and the Personal Genome Project (PGP), which tendency to create integrated and highly comprehensive human genome maps integrated with phenome data (22).
Application of computational modelling
Recently, the field of personalized medicine has been popularized with the fast development of the next generation sequencing, which caused higher throughput and lower costs (23,24). Additionally, public databases and platforms, i.e., GEO, TCGA, and ENCODE, contain significant amount of data for analysis (25). Systems biology with its multiomic data used in deep analyses for predictions, can provide additional insights into the mechanisms of complex diseases, especially for the various human cancers (26-28). Some recent developments regarding high-throughput technologies drive the systems biology in direction of creating more precise models to describe complex diseases. Mathematical, as well as computational models are used for the purposes to help us understand the omics data that are produced by high-throughput experimental techniques. The help of computational models in the area of systems biology, enables us to investigate the pathogenesis of complex diseases, further improve our understanding of latent molecular mechanisms, as well as promote treatment optimization strategies and new drug discoveries (29).
The bridge between theory and modelling in cancer can be accomplished by using two major complementary strategies—bottom-up approach, starting from the ‘omics’ data and top-down approach, where computer science and theoretical knowledge are the basis in creating models that describe dynamics of the system and its mechanisms. These two approaches can also be coupled to accomplish multi-scale models by combining wide biological scope and detailed mechanisms (30). In the context of translating cancer ‘omics’ data into clinical use, information sharing between medical research, epidemiology (cohort studies) and clinical medicine (prevention and treatment) is crucial. Multiscale models and computational platforms provide integration of the data and knowledge from these three areas.
Mathematical and computational modelling of biological systems at several scales is a good approach in discovering new drugs in clinical cancer therapy. At intracellular scale, these networks clarify how the cells regulate signaling or metabolic pathways in order to respond to the drug treatment or external perturbations (31). At intercellular scale, cell-cell communication networks explain how different cell types communicate using various ligands to speed up tumor growth, angiogenesis and metastasis (32). At tissue scale, studies on how these ligands diffuse and distribute in the 3D tumor space are quite valuable. With the advances of high-throughput technologies, systems biology rapidly develops. However, the development of mathematical modelling is constantly challenged by the new biological questions that arise (33).
Inhalation Drug Delivery Model—modelling of alveolar ducts in more detail
The Inhalation Drug Delivery Model is developed by our research group and because of that, we devote this part of the review paper to describe it in more detail. The results presented below are already published and we describe the modelling approach used by our research group in dealing with modelling of the described phenomena (34,35). Physicochemical characteristics of drug particles are important factors for the design of inhalation drug therapy, because they influence the distribution of drug delivery sites. That is the most essential determinant of where drug particles are delivered is the ventilation distribution. The reason for this is because drug particles, unlike respiratory gases (e.g, O2, CO2), are transported convectively with airflow in the lung airways.
Airflow is determined by the pressure difference (ΔP) between two points. In the case of the lung, along the airways the two points we should consider are the pressure at the airway opening (Pao) and the pressure at the alveoli (Palv). The distribution of inhaled air in the lungs is determined by the difference of time constant of each pathway (36). Pathway time constant is a product of airway resistance (R) and terminal compliance (C). Under normal breathing conditions (i.e., with spontaneous breathing frequencies), C normally dominates R (37), thus for normal/healthy subjects without tumors in the airways, the distribution of the terminal compliance C (i.e., downstream conditions) determines airflow distribution. Since the compliance of the lung is generally uniform (expect Capex < Cbase or gravity effects), the inspired air, and with it the inhaled drug particles, are considered to be distributed proportional to ventilation, namely, proportional to local lung size (e.g., the size of the lobe) (38). For instance, the larger lobe receives more air volume, thus more drug particles.
However, when airways are blocked by tumors, such as in lung cancer cases, the situation is quite different. Airflow distribution, thus the distribution of inhaled drug particles, is influenced by a combination of the local fluid mechanics around the tumors and the downstream flow conditions.
Flow conditions
Since the lungs consist of several hundred million alveoli (small air sacs where the gas exchange occurs), most of the lung volume is accounted by the alveolated region of the lungs. Although bronchioles and alveolated airways in the acinus contribute much less to airway flow resistance, compared to larger airways (bronchi) and the flow is nominally laminar, alveolar fluid mechanics are complex due to peculiar alveolated wall anatomy (which is also coupled with parenchyma tissue mechanics) and also depends on the exact location in the 8–9 generations along acinar tree.
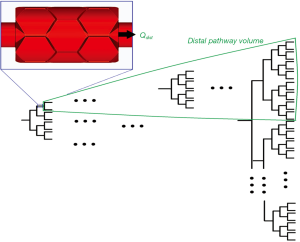
To determine the downstream flow conditions of airflow distribution (i.e., the distribution of inhaled drug particles) in the whole lung, we will sum alveolar flows of each path by calculating the alveolar flow of each generation of the path (Figure 1) in a 3-cell alveolated duct model (Figure 2) with various downstream boundary conditions (Qdist) (35), representing the distal volume to which that generation is connected (Figure 1, explained in detail below).
Detailed description is as follows. We assume that all lengths change with time in the same manner i.e., 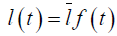 , where the overbar signifies the mean value. Hence, the volume
, where the overbar signifies the mean value. Hence, the volume 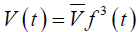 . The time rate of change of downstream distal volume produces the volume flow rate crossing the distal boundary of the model. Hence,
. The time rate of change of downstream distal volume produces the volume flow rate crossing the distal boundary of the model. Hence,
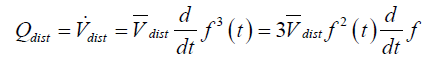
If we assume a sinusoidal variation of length with time; i.e.,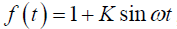 , where K=(ϕ–1)/(ϕ+1), ϕ=(1+C)1/3,
, where K=(ϕ–1)/(ϕ+1), ϕ=(1+C)1/3,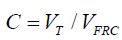 , VT is the tidal volume, VFRC is the functional residual capacity (volume), and ω=2π⁄T, where T is the breathing period, then
, VT is the tidal volume, VFRC is the functional residual capacity (volume), and ω=2π⁄T, where T is the breathing period, then 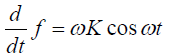 and
and 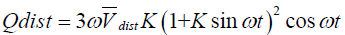 (39).
(39).
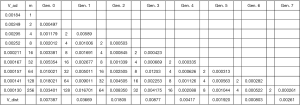
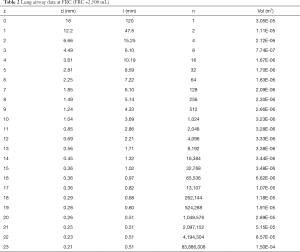
Distal volume numbers and average dimensions of alveolar ducts are given in Table 1. The variable f signifies the percentage of duct surface that is alveolated. In the table, both global, z, and, acinar, i, numbering of the airway generations are given. For instance, in the global system, the alveoli first appear in generation 15. This generation is designated generation 0 in the acinar system.
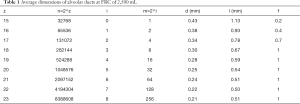
Full table
The total volume distal of a particular duct can be written as 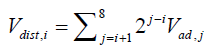 , Vad,i is the combined volume (alveolar volume surrounding the duct plus the duct volume) of a typical duct in acinar generation i. It is assumed that the alveolar volume surrounding a duct is proportional to the ratio of the duct’s alveolated surface area to the total surface area of all ducts in the acinus; i.e.,
, Vad,i is the combined volume (alveolar volume surrounding the duct plus the duct volume) of a typical duct in acinar generation i. It is assumed that the alveolar volume surrounding a duct is proportional to the ratio of the duct’s alveolated surface area to the total surface area of all ducts in the acinus; i.e., 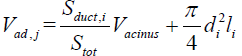 where Sduct,i=πdilifi,
where Sduct,i=πdilifi, 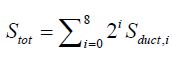 and
and 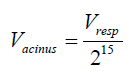 where
where 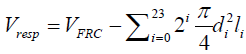 .
.
A sample calculation of Vdist is given below (Table 2) Vtot(0<z<23)=441.26ml, Vresp=2058.74ml, Vacinus=0.06283ml. It should be noted that the values given for Vdist are at FRC. These numbers can be adjusted to average values using
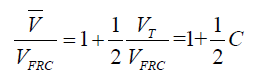
Full table
Data used to compute Vacinus are given in Table 2. The lengths and diameter data are from Weibel (40) and Weilbel et al. (41), scaled to FRC.
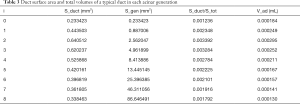
Total volumes (alveolar and duct) for a typical duct in each acinar generation are given in Table 3 (Stot). Components and total distal volume of a typical duct in each acinar generation are given in Figure 3.
Full table
Main limitations of the presented study are that this model is a parametric model, meaning it is simplified and not patient specific. However, the presented model is on an acinus level, so differences among people may not be noticeable. The possible easy adaptation of this model to different generations allows several parameters (i.e., flow) to be examined. In vivo tests are very hard to perform in animals, and even harder in humans due to regulations, so simulations can be helpful in this case to gain more knowledge on what happens on acinus level. This will be crucial in drug delivery analysis, in order to increase the efficiency of inhalers etc.
Finite element simulation in Lung Cancer Research
Success of any cancer therapy ultimately can be reduced to a degree of success of delivery of drug to the cancerous cells within tumor. From this physics point of view, the therapy can be considered as the transport problem of drug molecules, from blood vessels to the cell interior. On that path, drug molecules, and others important in the tumor formation and progression (as metabolic products, oxygen, etc.), go over different micro and macro environments—extracellular/intracellular space, interior of organelles, all of them in blood, as well as biological barriers—membranes of cells and organelles and walls of blood vessels. A lot of aspects of mass transport remains unknown, especially the biophysical mechanisms that govern the drug delivery.
The main research strategy counts on clinical investigations and laboratory, for example, those relying on nanotechnology (in view of transport called the oncophysics) (42-44). Paralelly, a great amount of efforts has been directed towards the development of computational tools for additional investigations of the intricate process of mass and transport exchange within capillary-tissue system in general and within tumors. As far as transport within tumors is considered, there exist additional complexities, mainly because of the variability of vessel diameters and lengths and irregular blood vessel branching (45,46). Experiments related to the flow within tumor vasculature unveiled that blood flow depends on several parameters including geometric resistance (47) (measure of network irregularities), viscous resistance (48), and RBC mechanical properties (49). Fundamental characteristics of blood flow inside tumor vasculature are presented in Jain (50), whilst the data regarding capillary wall transport parameters (hydraulic conductivity, vascular permeability and reflection coefficient) is shown in Jain (51).
Mass transport and exchange within cells is one of the most important processes in living organisms. The factors, which affect intracellular mass exchange, range from biochemical to mechanical, to signaling pathways (52). A computational framework for modeling mass exchange within cells, defined as ‘virtual cell’, is reported in Schaff et al. (53) and Moraru et al. (54), which was further used in many applications, as, for example in Slepchenko et al. (55).
Regarding computational methods, it can be stated that there is no general concept for mass transport within vessels (large and capillary blood vessels and lymph), extracellular space and cell interior. The basis for a general virtual lung transport model relies on recently developed Composite Smeared Finite Element (CSFE) (56), with improvements of accuracy (57), further enhanced to include lymphatic system (58), and generalized as a multiscale element which includes cell interior with organelles (59).
The CSFE brings fundamental change in the power of computational models in modeling complex process of mass transport in biological system, and particularly in drug delivery in tumors. This new methodology is general, robust and easy for implementation into modern software and tools within cloud computing. Data for the element include hydraulic, diffusive, electrical and chemical parameters for each physical domain and for biological barriers, and also volumetric fraction of each compartment. Physical fields are coupled by connectivity elements at each element node. A detailed study of accuracy of solutions obtained using the CSFE is given in Kojic (59).
Machine learning-based state-of-the-art methods for analysis of the NSCLC data
Many recent studies have analysed lung related TCGA data by using various machine learning approaches in order to predict quantity of interest. For instance, Yu et al. (60), applied several machine learning approaches on haematoxylin and eosin stained histopathology whole-slide lung adenocarcinoma images and squamous cell carcinoma patients from TCGA in order to differentiate between the shorter- and longer-term survivors with squamous cell carcinoma or stage I adenocarcinoma. For this purpose, authors extracted 9,879 quantitative image features and thereafter fed these features as inputs to elastic net-Cox proportional hazards models to identify the quantitative image features with most information, as well as to calculate survival indices. Thereafter, patients were into two categories—longer- or shorter-term survivors (based on their survival indices). In addition, authors used several machine-learning approaches, including naive Bayes, support vector machines and random forest, in order to: (I) distinguish malignancy from normal adjacent tissue, and (II) distinguish between adenocarcinoma and squamous cell carcinoma.
Adenocarcinoma and squamous cell carcinoma are the dominant histological types in lung cancers. Differentiating between these subtypes is very important as they have different treatment implications and prognosis. In Pineda et al. (61), authors used TCGA gene expression and DNA methylation data in order to discriminate between the two lung cancer subtypes. In this paper, authors used the ReliefF feature selection algorithm to determine relevant variables (genes), which were used to build a classification model based on naïve Bayes theorem. Authors confirmed biological relevance of their method by confirming that 93% of the selected genes are related to cancer.
Visual analysis of histopathology images with lung cell tissues is main method used by pathologists to determine the lung cancer stage, types and sub-types. In Coudray et al. (62), authors created a deep learning convolutional neural network based on histopathology images retrieved from TCGA in order to classify accurately the whole-slide pathology images into categories—normal lung tissue, adenocarcinoma or squamous cell carcinoma. Authors reported results that outperformed a human pathologist, with approximately 0.97 average AUC on a held-out population of whole-slide scans. In addition, authors used the neural network in prediction of the ten genes that are most commonly mutated in lung adenocarcinoma and discovered that six of the investigated genes—STK11, EGFR, FAT1, SETBP1, KRAS and TP53—can be prognosticated from pathology images (accuracy was in range from 0.733 to 0.856), as measured by the AUC on the held-out population.
In Yu et al. (63), authors exploited TCGA data of 538 lung adenocarcinoma patients to predict lung adenocarcinoma grade based on gene and protein expression levels. In addition, authors created an integrative histopathology-transcriptomics model in order to build the better prognostic (survival) predictions for stage I patients in comparison to the histopathology or gene expression studies alone.
Conclusions
Currently, researchers are focusing on the discovery of new drugs for cancer therapy, although molecular and cell biology had improved our understanding of many complex diseases in the last decades. In addition, research in lung cancer has become not only extensive but very much inter-professional, aiming to bring together important advances in various fields (medical and non-medical) for the sake of better understanding of the disease. This in turn can lead to optimized approaches in the diagnosis and treatment of each particular patient, which is usually seen as ultimate goal of “personalized medicine”. One such example is focused on predicting tumor growth. It can be used for multiple purposes such as providing information that can be used to disclose biological characteristics of the tumor (volume and cell kinetics), impact of various factors such as cell type, tumor size etc. These results could eventually lead to our better understanding of the natural history of the tumor that can influence our diagnostic and therapeutic decisions and tailor “optimal” treatment in lung cancer patients.
Acknowledgments
We acknowledgement efforts of Dr. Frank Henry in producing Figures 1-3 and Tables 1-3.
Funding: This work was suported by grants: III41007 and ON174028 Ministry of Education, Science and Technological Development of Serbia.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- World Health Organisation. Available online: https://www.iarc.fr/
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics. CA Cancer J Clin 2005;55:74-108. [Crossref] [PubMed]
- Vineis P, Alavanja M, Buffler P, et al. Tobacco and cancer: recent epidemiological evidence. J Natl Cancer Inst 2004;96:99-106. [Crossref] [PubMed]
- Darby S, Hill D, Auvinen A, et al. Radon in homes and risk of lung cancer: collaborative analysis of individual data from 13 European case-control studies. BMJ 2005;330:223. [Crossref] [PubMed]
- Krewski D, Lubin JH, Zielinski JM, et al. Residential radon and risk of lung cancer: a combined analysis of 7 North American case-control studies. Epidemiology 2005;16:137-45. [Crossref] [PubMed]
- Beveridge R, Pintos J, Parent MÉ, et al. Lung cancer risk associated with occupational exposure to nickel, chromium VI, and cadmium in two population‐based case–control studies in Montreal. Am J Ind Med 2010;53:476-85. [PubMed]
- Heck RM, Farrauto RJ, Gulati ST. Catalytic air pollution control: commercial technology. 3rd edition. Hoboken, New Jersey: John Wiley & Sons, 2009.
- Plon SE, Eccles DM, Easton D, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 2008;29:1282-91. [Crossref] [PubMed]
- Parkin DM, Boyd L, Walker L. The fraction of cancer attributable to lifestyle and environmental factors in the UK in 2010. Br J Cancer 2011;105:S77. [Crossref] [PubMed]
- Samet JM, Avila-Tang E, Boffetta P, et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res 2009;15:5626-45. [Crossref] [PubMed]
- Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol 2007;25:561-70. [Crossref] [PubMed]
- Requite project website. Available online: https://www.requite.eu/. Accessed 15.05.2019.
- LUNGCardRise project website. Available online: http://www.lungcardrise.eu/. Accessed 15.05.2019.
- Rastarget project website. Available online: https://cordis.europa.eu/project/rcn/108040/factsheet/en. Accessed 15.05.2019.
- Predect project website. Available online: https://www.imi.europa.eu/projects-results/project-factsheets/predect. Accessed 15.05.2019.
- Van der Heyden JH, Schaap MM, Kunst AE, et al. Socioeconomic inequalities in lung cancer mortality in 16 European populations. Lung cancer 2009;63:322-30. [Crossref] [PubMed]
- Balbin OA, Prensner JR, Sahu A, et al. Reconstructing targetable pathways in lung cancer by integrating diverse omics data. Nat Commun 2013;4:2617. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061. [Crossref] [PubMed]
- Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature 2010;464:993-8. [Crossref] [PubMed]
- Church GM. The personal genome project. Mol Syst Biol 2005;1:2005.0030.
- Lunshof JE, Bobe J, Aach J, et al. Personal genomes in progress: from the human genome project to the personal genome project. Dialogues Clin Neurosci 2010;12:47. [PubMed]
- Snyder M, Du J, Gerstein M. Personal genome sequencing: current approaches and challenges. Genes Dev 2010;24:423-31. [Crossref] [PubMed]
- Chen R, Snyder M. Promise of personalized omics to precision medicine. Wiley Interdiscip Rev Syst Biol Med 2013;5:73-82. [Crossref] [PubMed]
- Park C, Yu N, Choi I, et al. lncRNAtor: a comprehensive resource for functional investigation of long non-coding RNAs. Bioinformatics 2014;30:2480-5. [Crossref] [PubMed]
- Blanchet L, Smolinska A. Data fusion in metabolomics and proteomics for biomarker discovery. Statistical Analysis in Proteomics. New York, NY: Humana Press, 2016:209-23.
- Xu Y, Dou D, Ran X, et al. Integrative analysis of proteomics and metabolomics of anaphylactoid reaction induced by Xuesaitong injection. J Chromatogr A 2015;1416:103-11. [Crossref] [PubMed]
- Mayer P, Mayer B, Mayer G. Systems biology building a useful model from multiple markers and profiles. Nephrol Dial Transplant 2012;27:3995-4002. [Crossref] [PubMed]
- Ji Z, Xie Y, Guan Y. Involvement of P2X7 receptor in proliferation and migration of human glioma cells. Biomed Res Int 2018;2018:8591397. [Crossref] [PubMed]
- Stransky B, Barrera J, Ohno-Machado L, et al. Modeling cancer: integration of "omics" information in dynamic systems. J Bioinform Comput Biol 2007;5:977-86. [Crossref] [PubMed]
- Ji Z, Su J, Liu C, et al. Integrating genomics and proteomics data to predict drug effects using binary linear programming. PloS One 2014;9:e102798. [Crossref] [PubMed]
- Ji Z, Yan K, Li W, et al. Mathematical and computational modeling in complex biological systems. Biomed Res Int 2017;2017:5958321. [Crossref] [PubMed]
- Cheng F, Murray JL, Zhao J, et al. Systems biology-based investigation of cellular antiviral drug targets identified by gene-trap insertional mutagenesis. PLoS Comput Biol 2016;12:e1005074. [Crossref] [PubMed]
- Tsuda A, Henry FS, Butler JP. Gas and aerosol mixing in the acinus. Respir Physiol Neurobiol 2008;163:139-49. [Crossref] [PubMed]
- Henry FS, Tsuda A. Onset of alveolar recirculation in the developing lungs and its consequence on nanoparticle deposition in the pulmonary acinus. J Appl Physiol 2016;120:38-54. [Crossref] [PubMed]
- Otis AB, McKerrow CB, Bartlett RA, et al. Mechanical factors in distribution of pulmonary ventilation. J Appl Physiol 1956;8:427-43. [Crossref] [PubMed]
- Jaklitsch MT, Mery CM, Lukanich JM, et al. Sequential thoracic metastasectomy prolongs survival by re-establishing local control within the chest. J Thorac Cardiovasc Surg 2001;121:657-67. [Crossref] [PubMed]
- West JB. Distribution of blood and gas in lungs. Phys Med Biol 1966;11:357. [Crossref] [PubMed]
- Henry FS, Haber S, Haberthür D, et al. The simultaneous role of an alveolus as flow mixer and flow feeder for the deposition of inhaled submicron particles. J Biomech Eng 2012;134:121001. [Crossref] [PubMed]
- Weibel ER. A retrospective of lung morphometry: from 1963 to present. Am J Physiol Lung Cell Mol Physiol 2013;305:L405-8. [Crossref] [PubMed]
- Weibel ER, Sapoval B, Filoche M. Design of peripheral airways for efficient gas exchange. Respir Physiol Neurobiol 2005;148:3-21. [Crossref] [PubMed]
- Ferrari M. Frontiers in cancer nanomedicine: directing mass transport through biological barriers. Trends Biotechnol 2010;28:181-8. [Crossref] [PubMed]
- Koay EJ, Ferrari M. Transport Oncophysics in silico, in vitro, and in vivo. Phys Biol 2014;11:060201. [Crossref] [PubMed]
- Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol 2015;33:941. [Crossref] [PubMed]
- Less JR, Skalak TC, Sevick EM, et al. Microvascular architecture in a mammary carcinoma: branching patterns and vessel dimensions. Cancer Res 1991;51:265-73. [PubMed]
- Roberts WG, Palade GE. Neovasculature induced by vascular endothelial growth factor is fenestrated. Cancer Res 1997;57:765-72. [PubMed]
- Sevick EM, Jain RK. Geometric resistance to blood flow in solid tumors perfused ex vivo: effects of tumor size and perfusion pressure. Cancer Res 1989;49:3506-12. [PubMed]
- Sevick EM, Jain RK. Viscous resistance to blood flow in solid tumors: effect of hematocrit on intratumor blood viscosity. Cancer Res 1989;49:3513-9. [PubMed]
- Sevick EM, Jain RK. Effect of red blood cell rigidity on tumor blood flow: increase in viscous resistance during hyperglycemia. Cancer Res 1991;51:2727-30. [PubMed]
- Jain RK. Determinants of tumor blood flow: a review. Cancer Res 1988;48:2641-58. [PubMed]
- Jain RK. Transport of molecules across tumor vasculature. Cancer Metastasis Rev 1987;6:559-93. [Crossref] [PubMed]
- Rangamani P, Iyengar R. Modelling spatio-temporal interactions within the cell. J Biosci 2007;32:157-67. [Crossref] [PubMed]
- Schaff J, Fink CC, Slepchenko B, et al. A general computational framework for modeling cellular structure and function. Biophys J 1997;73:1135-46. [Crossref] [PubMed]
- Moraru II, Schaff JC, Slepchenko BM, et al. Virtual Cell modelling and simulation software environment. Iet Syst Biol 2008;2:352-62. [Crossref] [PubMed]
- Slepchenko BM, Schaff JC, Macara I, et al. Quantitative cell biology with the Virtual Cell. Trends Cell Biol 2003;13:570-6. [Crossref] [PubMed]
- Kojic M, Milosevic M, Simic V, et al. A composite smeared finite element for mass transport in capillary systems and biological tissue. Comput Methods Appl Mech Eng 2017;324:413-37. [Crossref] [PubMed]
- Milosevic M, Simic V, Milicevic B, et al. Correction function for accuracy improvement of the Composite Smeared Finite Element for diffusive transport in biological tissue systems. Comput Methods Appl Mech Eng 2018;338:97-116. [Crossref] [PubMed]
- Kojic M, Milosevic M, Simic V, et al. Extension of the composite smeared finite element (CSFE) to include lymphatic system in modeling mass transport in capillary systems and biological tissue. J Serbian Soc Comput Mech 2017;11:108. [Crossref] [PubMed]
- Kojic M. Smeared Concept as a General Methodology in Finite Element Modeling of Physical Fields and Mechanical Problems in Composite Media. J Ser Soc Comp Mech 2018;12:1-16. [Crossref]
- Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016;7:12474. [Crossref] [PubMed]
- Pineda AL, Ogoe HA, Balasubramanian JB, et al. On Predicting lung cancer subtypes using ‘omic’ data from tumor and tumor-adjacent histologically-normal tissue. BMC cancer 2016;16:184. [Crossref] [PubMed]
- Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559. [Crossref] [PubMed]
- Yu KH, Berry GJ, Rubin DL, et al. Association of omics features with histopathology patterns in lung adenocarcinoma. Cell Syst 2017;5:620-7.e3. [Crossref] [PubMed]
Cite this article as: Filipovic N, Sustersic T, Vulovic A, Tsuda A. Big Data and machine learning: new frontier in lung cancer care. Shanghai Chest 2019;3:51.