
Lung cancer histology-driven strategic therapeutic approaches
Introduction
Among all lung primary pulmonary carcinomas, non-small cell lung cancer (NSCLC) accounts for about 75–80% of all cases of pulmonary malignancies (1) and radical surgery still remains the only treatment with curative intent. Different factors influence the best management of patients with NSCLC, including those associated with the patient (respiratory functions, performance status, past medical history) and/or related to the tumour characteristics (stage, molecular predictive biomarkers), including histology (or histological type) (2,3). Of note, epidemiologic works revealed a significant increase in the incidence of adenocarcinomas, particularly in women, from 1979 to 1998 (4,5), possibly due to smoking habit and a different cigarettes composition leading to greater exposure of cigarette carcinogens in the peripheral alveolar regions of the lungs (6).
In regards with histological tumour definition, clinicians generally subdivide lung cancer into two major groups: small cell lung cancer (SCLC) and NSCLC (7). This dichotomic classification has been considered sufficiently exhaustive for the management of patients with lung cancer up to the introduction of chemotherapy regimen with pemetrexed and/or bevacizumab, requiring the NSCLC subcategorization at least into squamous versus non-squamous cell carcinoma (8-11). In the meantime, the huge amount of molecular information derived from gene expression profiling and next generation sequencing studies evidenced several genetic alterations in lung cancer oncogenic drivers predicting efficacy using specific targeted molecules (small inhibitors and monoclonal antibodies), somehow limiting the role of tumour histotype (12-16).
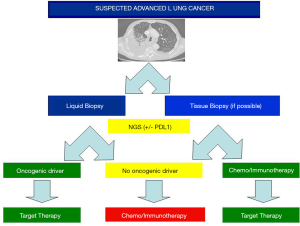
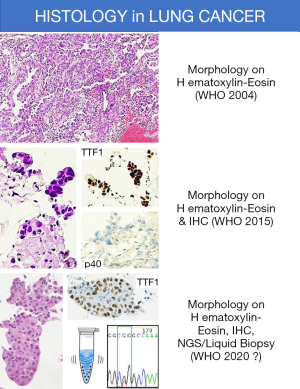
Indeed, in the last decade the most important and effective changes in the therapeutic approach of patients with lung cancer are related to the discovery of molecular impairments in druggable oncogenic drivers and immunotherapy using humanized monoclonal antibodies blocking the programmed death (PD)-1/PD-ligand (PD-L)-1 checkpoint (12). In addition, the advent of liquid biopsy already permits to evidence key genetic alterations using highly sensitive methods to sequence DNA and RNA (13), preventing to obtain tissue samples using invasive procedures and facilitating the introduction of ongoing drugs against histology-agnostic genetic alterations (e.g., NTRKs) (Figure 1) (14-16). All these features are deeply challenging the role of classic lung tumor histologic definition and enrich the meaning of histology with additional molecular information (Figure 2).
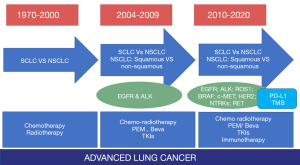
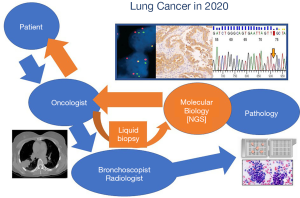
Histology is no more limited to morphology
Histologic definition of lung cancer is based on criteria posed by the most recent 2015 WHO classification (17). Since more than two third of NSCLC are not resectable at diagnosis, histologic features derived from examination of surgically-excised tumors are not always translatable in cytology/small biopsy. In addition, limitation of histology-based therapies relies in the simple distinction between squamous cell versus non-squamous cell carcinoma as the mainstay for clinical management of patients with NSCLC.
Although recommended for NSCLC subtyping, the helpful role of immunohistochemistry in supporting morphology is somehow limited by the “aberrant” expression of some biomarkers (e.g., p63 reactivity in adenocarcinoma or CK7 in squamous cell carcinoma) (Table 1). In this setting, the best single marker for adenocarcinoma is TTF1 (clone 8G7G3/1) and the best single marker for squamous cell carcinoma is p40, realizing a two-hit, sparing material algorithm suitable for both cytology and small biopsy specimens (Figure 3) (Table 2) (18-24).
Table 1
| Antibody | ADC | SQC | SCLC/LCNEC | Carcinoid tumors | Metastasis |
|---|---|---|---|---|---|
| TTF-1 | ~80% | Never (clone 8G7G3/3) | 60–80% | Peripheral type | Endometrial cancer# |
| Napsin | ~80% | Never | Never | Never | some renal cancer |
| CK7 | Almost all | 30–60% | >50% | >50% | several other primaries |
| p63 | 30% | Almost all | Almost never | Never | SQC from other sites urothelial carcinoma |
| p40 | Almost never° | Almost all | Almost never | Never | SQC from other sites urothelial carcinoma |
| CK5/6 | 10% | Almost all | Almost never | Never | SQC from other sites urothelial carcinoma |
| CDX2 | Enteric, colloid | Never | 10–20% | Never | Colo-rectal ADC, ADC with enteric differentiation |
| CK20 | Enteric, colloid, mucinous | Never | Never | Never | ADC with intestinal differentiation |
| ER/PgR | 10–20%* | Never | Almost never | Sometimes | Breast, GYN tract |
#, TTF-1 clones SPT24 and SP141 may be expressed in several extra-pulmonary tumors (e.g., breast and gastric cancer, mesothelioma); *, ER/PgR are mainly expressed at low intensity in well-differentiated adenocarcinoma arising in women; °, p40 is focally expressed in a small subset of lung adenocarcinoma. ADC, adenocarcinoma; SQC, squamous cell carcinoma; SCLC/LCNEC, small cell lung cancer/large cell neuroendocrine carcinoma; TTF-1, thyroid transcription factor 1; CK, cytokeratin; CDX2, caudal type homeobox 2; GYN, gynaecologic tract; GI, gastrointestinal tract; CK, cytokeratin; ER, estrogen receptor; PgR, progesterone receptor.
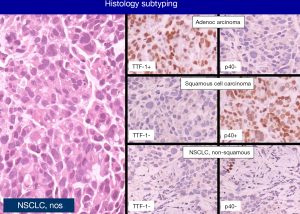
Table 2
| Histology | First choice | Second choice |
|---|---|---|
| Adenocarcinoma | TTF-1 (clone 8G7G3/1) | Napsin; CK7 |
| Squamous cell carcinoma | p40 | p63; CK5/6; desmocollin |
| NE tumors | chromogranin; synaptophysin | CD56 |
| Others | NUT (NUT-carcinoma) | |
| CDX2 (adenocarcinoma, enteric and colloid variants) | ||
| SMARCA4 (undifferentiated SMARCA4-deficient carcinoma) | ||
NE, neuroendocrine.
The list of primary lung cancers identified in the WHO classification comprises 3 main categories (adenocarcinoma, squamous cell carcinoma, neuroendocrine tumours) and poorly-differentiated or undifferentiated tumours lacking any differentiation at light microscope analysis or extensive immunostaining [e.g., large cell carcinoma (LCC) and sarcomatoid carcinoma].
Limitations of conventional histologic examination to disclose the original cell differentiation in poorly-differentiated/undifferentiated carcinomas are well-demonstrated by recent molecular studies in large cell neuroendocrine carcinoma (LCNEC), undifferentiated LCC [namely NSCLC, not otherwise specified (n.o.s.) on cytology and small biopsy] and sarcomatoid carcinoma.
LCNEC has been considered a variant of LCC in the previous 2004 WHO classification (2) of lung cancer and then included as poorly-differentiated carcinoma into the rubric of neuroendocrine tumours in the last classification (17). The diagnosis requires a clear-cut neuroendocrine differentiation at morphology and at the immunohistochemistry level. The prognosis is quite similar to that of SCLC (17). However, the overall survival at 5 years reported in literature is ranging from 13% to 51% in stage I, consistently suggesting that this challenging diagnosis depends on the application of rigorous pathologic criteria (25).
Indeed, next-generation sequencing (NGS) molecular analysis demonstrates that LCNEC is a biologically heterogeneous basket of tumours segregated in at least 3 main subgroups: SCLC-like (TP53+RB1 and MYCL amplification), NSCLC-like (lack of co-altered TP53+RB1 and mutations of STK11, KRAS, KEAP1) and carcinoid-like (MEN1 mutations and low mutation burden). SCLC-like showed higher proliferative activity than NSCLC-like tumours (P<0.0001), while NSCLC-like LCNEC harboured distinctive genomic alterations, including mutations of NOTCH family genes regulating neuroendocrine differentiation (26).
In addition, Derks et al. (27) showed that LCNEC carrying a wild-type RB1 gene or expressing the RB1 protein do better with NSCLC-like treatment (gemcitabine/Taxol) than with SCLC-chemotherapy (etoposide), confirming previous experiences (28).
Karlsson et al. (29) investigated LCCs with (n=32) or without (n=41) neuroendocrine features using massive parallel sequencing for mutations in 26 cancer-related genes and gene fusions in ALK, RET, and ROS1. Based on immunostains, LCC without NE differentiation were subdivided in adenocarcinoma-like (TTF1/napsin +), squamous-like (CK5/p40) and “null” type. The most common alterations in LCC lacking NE features were TP53 (83%), KRAS (22%), MET (12%) mutations, while TP53 (88%), STK11 (16%), and PTEN (13%) mutations were significantly higher in LCNEC, demonstrating that LCC with and without NE features follow different molecular pathways impacting in therapeutic decisions.
LCC is a non-small-cell carcinoma lacking morphologic differentiation of either adenocarcinoma or squamous cell carcinoma, basically representing a highly aggressive tumour with an end-stage cell differentiation (30,31). Rekhtman et al. (32) analysed 102 LCC with immunostaining (TTF-1 versus p40) and molecular gene alterations (EGFR, KRAS, BRAF, MAP2K1/MEK1, NRAS, ERBB2/HER2 mutations and ALK rearrangements versus PIK3CA and AKT1 mutations) for adenocarcinoma and squamous cell carcinoma, respectively. Of note, molecular alterations characteristic of adenocarcinoma occurred in tumours with immunoprofiles of adenocarcinoma or marker-null, but not in tumours with squamous immunoprofile (combined mutation rate 50% vs. 30% vs. 0%, respectively; P<0.001), whereas the sole PIK3CA mutation occurred in a tumor with squamous profile (5%). Then, the majority (80%) of LCC did represent poorly-differentiated forms of adenocarcinoma or squamous cell carcinoma.
Similarly, Driver et al. (33) reclassified 17 LCC as adenocarcinoma (9 cases with mutations in KRAS, EGFR, BRAF) and 8 as squamous cell carcinoma (PIK3CA, CDKN2A mutations) using NGS technology. Pelosi et al. (34) dissected 30 LCC by unsupervised targeted next generation sequencing analysis demonstrating that 3 cases showing TTF1-/p40+ phenotype harboured TP53 only in keeping with a squamous cell lineage, while the others 90% featuring various phenotypical combinations of TTF1 and p40 comprised ATM, BRAF, CDKN2A, EGFR, ERBB4, FBXW7, FLT3, KRAS, NRAS, PIK3CA, PTPN11, RET, SMAD4, SMO, STK11, or TP53 mutations in keeping with adenocarcinoma lineage. Another study investigating lineage-specific immunomarkers, EGFR and KRAS mutations and ALK rearrangement in 121 LCC along the spectrum of variants provided by the 2004 WHO classification evidenced that all 47 LCNEC had a true neuroendocrine cell lineage without gene alterations, whereas all 24 basaloid and 2 lymphoepithelioma-like carcinomas showed squamous cell markers (35). Eighteen out of 22 clear cell carcinomas had glandular differentiation, with KRAS mutations in 39% of cases. Eighteen out of 20 undifferentiated LCC showed glandular differentiation upon immunohistochemistry, with exon 21 L858R EGFR mutation in one (5%) tumour, exon 2 KRAS mutation in eight (40%) tumours, and ALK translocation in one (5%). All 6 LCC of rhabdoid type expressed TTF-1 and/or CK7, 50% of which also harboured KRAS mutations (35). At the end, molecular alterations were restricted to LCC having an adenocarcinoma cell differentiation and stratification of LCC using immunohistochemistry and molecular analysis revealed a direct correlation between phenotypic and genotypic arrangements.
Sarcomatoid carcinoma is an umbrella term to indicate a group of poorly-differentiated/undifferentiated NSCLC showing sarcoma-like (giant and/or spindle cell component) or true sarcomatous (mainly chondrosarcoma, osteosarcoma and rhabdomyosarcoma) differentiation with or without a component of conventional NSCLC (17,36), then including different variants (pleomorphic, spindle cell, giant cell carcinomas, carcinosarcoma and pulmonary blastoma). Previous studies on relatively large series demonstrated that at immunohistochemistry thyroid transcription factor-1 (TTF-1) and cytokeratin 7 were positive in 55% and 70% of spindle and/or giant cell carcinomas and 43% and 63% in pleomorphic carcinomas, supporting the metaplastic histogenetic theory for these tumours, ancestrally starting from a conventional histology possessing a genetic EMT program (37). Even at molecular level, sarcomatoid carcinomas harbour mutations involving oncogenic drivers of adenocarcinoma or squamous cell carcinomas, including KRAS, EGFR, TP53, STK11, NOTCH1, NRAS, PI3KCA and BRAF (38-40). A significantly higher rate of c-MET skipping mutations in exon 14 and MET amplification have been reported in this rare histology (41,42).
Finally, histology has also a role in explaining primary or secondary resistance to tyrosine kinase inhibitors (TKI) in tumours harbouring mutations or rearrangements in the most common oncogenic drivers, but also in chemotherapy and immunotherapy (43-67). Indeed, a histologic change from adenocarcinoma to small cell or squamous cell carcinoma have been well-demonstrated in about 10% of EGFR mutated or ALK rearranged lung cancer (68,69).
More rarely, a sarcomatoid “transformation” due to activation of EMT has been described by Hsieh et al. (48) in 6 cases of adenocarcinoma (5 EGFR mutated, 1 ROS1 rearranged). Histologic change to sarcomatoid carcinoma in TKI resistant adenocarcinomas is accompanied by PD-L1 over-expression and c-MET gene alterations.
The pass to sarcomatoid histology has been previously demonstrated in EGFR-mutated cell lines of lung adenocarcinoma, and concurrent acquisition of other gene alterations (i.e., T790M EGFR or NKx2-4 mutation) was recently observed (49-52).
Histologic “transformation” has clearly a key therapeutic impact in predicting the urgent need for alternative therapies in NSCLC progressing on TKI.
Histology-based chemotherapy
While multivariate analyses on large cooperative groups on chemotherapy in advanced NSCLC stated that histotype is not a determinant factor of efficacy and have a little, if none, prognostic significance (7), alternative chemotherapeutic protocols have reported a different response rate in regards with lung cancer histology.
Tegafur-uracil in adjuvant setting improved overall survival of Japanese patients with stage I adenocarcinoma subtype (70). Again, Georgoulias et al. (71) revealed that the regimen including gemcitabine + docetaxel was significantly more effective in patients with adenocarcinoma histology than in non-adenocarcinomatous NSCLC. On the other hand, patients with non-adenocarcinoma NSCLC had a significant better response to cisplatinum + docetaxel than those with adenocarcinoma histology, then stating that histological type had an important predictive role at univariate and multivariate statistical analysis (71).
Another controversial lung tumour entity lacking standard chemotherapeutic protocols is LCNEC. As originally described by Travis et al. (72), LCNEC seems to have a very poor outcome, quite similar to SCLC with which LCNEC shares various genetic alterations (17,73). Recent retrospective studies have demonstrated a significant higher survival in patients receiving a SCLC-like chemotherapy protocol (platinum + etoposide/VP16) either in adjuvant and metastatic settings (28,74,75). In addition, a prospective study of adjuvant chemotherapy for pulmonary LCNEC by Iyoda et al. (75) confirmed that patients (n=15) undergoing cisplatin and VP16 after surgery had a significant survival improvement at 2 and 5 years. These results were subsequently confirmed in other works (26-28), particularly when LCNEC had SCLC-type alterations, namely TP53+RB1 co-mutation/loss (26).
The key role of histologic subtyping in the best choice of chemotherapeutic protocols has recently merged from a planned subgroup analysis of the JMEN phase III clinical trial comparing pemetrexed and gemcitabine in association with platinum. More in details, Scagliotti et al. (11) reported a survival advantage for cisplatin + pemetrexed over cisplatin + gemcitabine (11.8 versus 10.4 months) in patients with non-squamous NSCLC, with a more impressive result in patients with adenocarcinoma (12.6 versus 10.9 months; P=0.03). Of note, the analysis in the population with NSCLC n.o.s. failed to reveal a significant difference in survival. Based on this trial, histological subtyping was considered mandatory in planning a regimen with cisplatin + pemetrexed in chemonaive patients with non-squamous NSCLC, lacking targetable genetic alterations and/or not amenable to immunotherapy.
Nevertheless, another phase III study enrolling 436 patients did not demonstrate significant differences for quality of life and in overall survival between the two treatment arms (pemetrexed/carboplatin, 7.3 months versus gemcitabine/carboplatin, 7.0 months; P=0.63) with less hematologic toxicity and less need for supportive care (76).
In a phase II trial (10), 99 patients randomly assigned to bevacizumab or plus carboplatin and paclitaxel or carboplatin and paclitaxel alone, treatment with carboplatin and paclitaxel plus bevacizumab resulted in a higher response rate (31.5% vs. 18.8%), longer median time to progression (7.4 vs. 4.2 months) and a modest increase in survival (17.7 vs. 14.9 months). Bleeding was the most prominent adverse event and was major haemoptysis was associated with squamous cell histology, tumor necrosis and cavitation, and disease location close to major blood vessels. Then, patients with non-squamous cell histology appeared to represent a subset with improved outcome and acceptable safety risks. Based on this previous study, a randomized study by the Eastern Cooperative Oncology Group (ECOG) including 878 patients with recurrent or advanced non-small-cell lung cancer (stage IIIB or IV) compared chemotherapy with paclitaxel and carboplatin alone versus paclitaxel and carboplatin plus bevacizumab was designed, but squamous-cell carcinoma histology was a major parameter in excluding patients for enrolment (77).
So, non-squamous carcinoma histology has become a selective factor when using chemotherapeutic regimens comprising pemetrexed or bevacizumab (78) (Table 3).
Table 3
| Agent | Predictive factor |
|---|---|
| Bevacizumab | Histology (non-squamous) |
| Pemetrexed | Histology (non-squamous) |
| EGFR inhibitors | EGFR mutation |
| ALK inhibitors | ALK rearrangement |
| ROS1 inhibitors | ROS1 rearrangement |
| BRAF inhibitors | BRAF (V600E) mutation |
| PD-1/PD-L1 blockers | PD-L1 ≥50% (first line with pembrolizumab) |
| NTRK inhibitors | NTRKs rearrangements |
NSCLC, non-small cell lung cancer.
Histology, molecular biology and targeted therapy
Several lung cancer oncogenic drivers acting even as targetable genes are specifically related to adenocarcinoma histology, namely EGFR, KRAS, BRAF, HER2, c-MET mutations and ALK, ROS1, RET or NTRKs rearrangements (79).
In some way, these molecular alterations intrinsically possess a diagnostic value. In other words, the finding of the aforementioned genetic abnormalities consistently indicates that the analysed tumour has an adenocarcinoma histology, although not in an absolute meaning (80-84) (Figure 4).
Indeed, sporadic reports have described cases of squamous cell carcinoma, SCLC/LCNEC, sarcomatoid carcinoma harbouring EGFR mutations or ALK and ROS1 rearrangements (85-87). Interestingly, clinical response to TKI in non-adenocarcinoma lung cancer with EGFR sensitive mutations is significantly lower than that observed in mutated adenocarcinomas and similar to chemotherapy (87).
Nevertheless, Cooperative Groups (88) have prospectively performed a genome-based diagnosis testing 5,145 lung cancer of different histology, successfully assigning a histopathologic type in 75% of cases. EGFR, KRAS, ERBB2, BRAF, STK11, ALK gene alterations and NKX2-1 amplification were significantly restricted to adenocarcinoma, whereas DDR2, FGFR3, NFE2L2 mutations and SOX2 or FGFR1 amplification in squamous cell carcinoma. MYCN amplification and RB deletion occurred in SCLC. These important results open to a molecular diagnosis providing a genetic diagnosis/histology and oncogenic driver alterations permitting specific tailored therapies.
In this revolutionary landscape dominated by identification of molecular gene alterations and specifically-related pharmaceutical agents using multigene panel by NGS broadly covering all types of lung cancer in routine practice, histology is losing its importance even in selecting molecular tests (89,90).
Indeed, molecular determinations can already override histology in poorly-differentiated NSCLC resulting particularly challenging at light microscopy and immunohistochemistry (e.g., adenocarcinomas with “squamoid” pattern, adenosquamous carcinoma or squamous cell carcinoma with pseudoglandular differentiation), finally revealing gene alterations specifically characterizing lung cancer histotypes (e.g., EGFR or KRAS mutations in adenocarcinoma or PI3KCA and miR-205 in squamous cell carcinoma) concomitantly having a therapeutic significance (91,92).
Several studies investigating gene expression profiles and massively sequencing tumor DNA have proposed a molecular classification of lung cancer secondary to evidence of histology-specific genetic alterations, then improving the diagnosis and treatments particularly in adenocarcinoma.
The seminal study by Bhattacharjee et al. (93) analyzed mRNA expression levels in 139 resected adenocarcinomas clustering expression data in distinct subclasses, evidencing 1 subset with high relative expression of neuroendocrine genes and poor prognosis and demonstrating the ability to discriminate primary lung adenocarcinomas from metastases of extra-pulmonary origin.
In comparison to conventional histologic classification, the added value of these studies is in highlighting the genetic complexity of lung cancer. In other words, dealing with a common squamous cell carcinoma, molecular analysis may reveal the hidden heterogeneity stratifying the tumor in different biologic entities. Wilkerson et al. (94) clustered squamous cell carcinomas at mRNA expression in at least 4 subtypes, named primitive, classical, secretory, and basal demonstrating the worst survival outcome of the primitive type (P<0.05) and the relationship between the different profiles and biological processes (primitive: proliferation; classical: xenobiotic metabolism; secretory: immune response; basal: cell adhesion).
Despite the literature clearly indicates that molecular alterations involving EGFR, KRAS, BRAF, HER2 and ALK or ROS1 rearrangements are basically detected in adenocarcinoma only or in combined adenocarcinoma (80,95-105), no study has been finalized at demonstrating the diagnostic role of mutational analysis in differentiating lung cancer histologic types.
Several studies have identified genomic alterations and actionable mutations in lung adenocarcinoma and even in squamous cell carcinoma and SCLC (106-109) demonstrating that just a limited number of somatically mutated genes overlap all three subtypes and possibly permitting the use of a set of genes to address lung cancer diagnosis.
The increase incidence of adenocarcinoma histotype among all primary lung cancers may more likely lead to multiple lesions in the lungs. The distinction between different primary tumors or intrapulmonary metastases has a fundamental clinical value influencing the diagnosis, the tumor stage and then the patient management. Since it is not always reliable to distinguish separate primary lung cancers from intrapulmonary metastasis on histology and immunostains, Chang et al. (110) investigated the value of molecular analysis by NGS in 76 tumor pairs from 60 patients. NGS classified tumor pairs into 51 definite separate primary cancers and 25 metastatic tumors evidencing discordant results with histology in 17 cases (22%), particularly in metastatic cancers (44% discordant). These results robustly support the diagnostic role of NGS to assist conventional histology and immunohistochemistry in defining primary versus metastatic multiple lung cancer in clinical practice.
Molecular histology in liquid biopsy
The diagnosis of lung cancer is generally based on identification of tumor cells at light microscope examination, but the ever increasing need to acquire tumor tissue to deeply investigate molecular profile in naïve NSCLC, the mechanisms underlying secondary resistance to TKI during disease progression and intratumor heterogeneity is partially hampered by the minimal availability of re-biopsy (111-117).
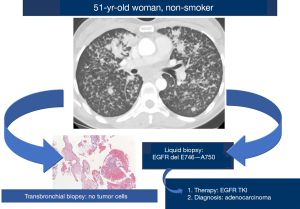
In the era of targeted treatments guided by recognition of “druggable” oncogenic drivers, the advent of liquid biopsy providing a comprehensive genetic profile of lung cancer through analysis of circulating tumor DNA (ctDNA), circulating tumor cells or exosomes is a revolutionary approach over conventional sampling procedures (118,119). Liquid biopsy is recommended in the new College of American Pathologists (CAP)/International Association for the Study of Lung Cancer (IASLC)/Association for Molecular Pathology (AMP) guideline for molecular testing of patients with NSCLC (113).
According to the statement paper from the IASLC, Rolfo et al. (120) proposed the use of liquid biopsy (circulating cell-free tumor DNA in plasma) in case of biopsy containing insufficient tumor tissue or when tissue specimens are not obtainable by traditional procedures (suboptimal clinical condition of the patients, unfavorable tumor site, high risk of major complications). Tissue biopsy is generally more expensive than a blood draw, particularly when repeated analyses are required during disease progression on targeted therapy. In addition, liquid biopsy has a shorter turnaround time and is more representative of the entire biology of metastatic NSCLC.
Zhang et al. (121) analyzed genetic alterations of 48 tissue biopsy and matched liquid biopsy from early-stage NSCLC using 546 genes capture-based NGS assay demonstrating a concordant setup, particularly in squamous cell carcinoma. Then, a model of 14 gene mutations (TP53, SLIT2, NOTCH3, MTOR, LIFR, MRE11A, AID2, ERCC3, KCNH2, CDC25C, RB1, ALK, NFE2L2, FBXW7) showed an overall accuracy of 90% in the training and testing set aimed at histologic subtyping.
In previous studies, plasma NGS (probe set including KRAS, EGFR, ALK, HER2, BRAF, NRAS, PI3K3CA, MET, MEK1, TP53 mutations and ALK, ROS1, RET rearrangements) performed in progressive NSCLC was able to identify gene mutations, amplification and rearrangements with a specificity of 100% and a sensitivity of 77% when compared with tumor tissue genotype (122). In addition, plasma NGS permitted to recognized actionable gene alterations (EGFR mutation and MET amplification) in patients with incomplete tissue genotyping without false positive results.
In another study, Müller et al. (123) investigated a cohort of 82 patients with non-squamous NSCLC by a massively parallel sequencing liquid biopsy assay covering 39 genes (NEOliquid) matching plasma and tissue samples. The concordance was 98%, the sensitivity 70% and the specificity 100%. Among discordant cases, some cases had a driver mutation only in plasma (IDH1, RET, MET mutations).
More recently, Aggarwal et al. (13) prospectively enrolled 323 patients with metastatic NSCLC routinely tested with plasma NGS using a 73-gene commercial platform. Among 94 patients with plasma NGS alone there were 31 targetable mutations (33%), while the 229 patients with plasma and tissue NGS, an actionable mutation was disclosed in 47 tissue biopsy (20.5%) and in 82 liquid biopsies (35.8%). The authors recommended an integration of plasma NGS testing into the routine practice, possibly increasing the spectrum of targetable mutations if compared with conventional tissue biopsy.
Finally, Zhou et al. (124) compared tumor DNA and cell-free DNA in 63 patients diagnosed as LCNEC by target-capture sequencing demonstrating that the mutation concordance was 90% and patients with LCNEC presenting a SCLC-like genomic subtyping (mutations or copy number loss in both RB1 and TP53) have a shorter overall survival and a superior response to etoposide-platinum chemotherapy.
Although liquid biopsy is still questionable in terms of sensitivity and not recommended to replace a diagnostic tissue biopsy, the aforementioned works consistently challenge this dogma (Figure 5).
Histology-agnostic therapy
Recently, the U.S. Food and Drug Administration has approved 2 drugs, pembrolizumab and larotrectinib, in a histology-agnostic setting. In other words, innovative clinical trials seem to favor the identification of targetable molecular alterations over conventional histology determining the tumor primary and histological tumor subtype (14,15,125).
Pembrolizumab, an anti-programmed cell death-1 (PD-1) monoclonal antibody (mAb), has received accelerated approval for the treatment of adult and pediatric patients with unresectable or metastatic solid tumors harboring microsatellite instability-high or deficient DNA mismatch repair. Similarly, nivolumab, another anti-PD-1 mAb, experienced an accelerated approval for adult and pediatric patients with microsatellite instability-high or deficient DNA mismatch repair metastatic colorectal cancer progressed after standard chemotherapy. Finally, larotrectinib, an oral and selective inhibitor of tropomyosin receptor kinases (TRK), demonstrated unprecedented efficacy on unresectable or metastatic solid tumors with neurotrophic tropomyosin receptor kinase (NTRK)-fusion proteins in adult and pediatric patients (14,15,125).
All these data represent a novel and revolutionary approach to cancer treatment based on biomarker biomarker-selected patients and characterized by high clinical efficacy, durable response and unselected patients’ population.
In other words, agnostic-histology development model of clinical trials together with the increasing accessibility to high-throughput genetic analysis (e.g., NGS) (126) and minimally invasive liquid biopsy could re-design the future role of conventional tissue biopsy and pathologists involved in oncology.
Conclusions
Despite the great efforts by an international panel of expert pulmonary pathologists of the WHO/IASLC in periodically developing new classification of lung tumours characterized by a good reproducibility and simplicity as well as clinical relevance, conventional histology will have a limited role in the future management of patients with lung cancer.
The concept of histology (or tumour histologic type) significantly changed in the last years with the overbearing entrance of molecular information. Nowadays, the modern meaning of histology should incorporate key genetic information permitting a more precise diagnosis, a correct tumour stage in pulmonary multiple cancers and molecularly guided targeted therapies (Figure 6).
The possibility to obtain robust histotype-related multigene data from liquid biopsy could partially replace the need for tumour tissue, dramatically introducing a novel and non-invasive paradigm in approaching patients with lung cancer.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/shc.2020.01.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Travis WD, Brambilla E, Muller-Hermelink HK, et al. editors. Tumours of the Lung, Pleura, Thymus and Heart. Pathology & Genetics. World Health Organization Classification of Tumours. IARC Press, Lyon, 2004.
- Birim O, Kappetein AP, van Klaveren RJ, et al. Prognostic factors in non-small cell lung cancer surgery. Eur J Surg Oncol 2006;32:12-23. [Crossref] [PubMed]
- Travis WD, Lubin J, Ries L, et al. United States lung carcinoma incidence trends: eclining for most histologic types among males, increasing among females. Cancer 1996;77:2464-70. [Crossref] [PubMed]
- Tyczynski JE, Bray F, Parkin DM. Lung cancer in Europe in 2000: epidemiology, prevention, and early detection. Lancet Oncol 2003;4:45-55. [Crossref] [PubMed]
- Boffetta P, Pershagen G, Jockel KH, et al. Cigar and pipe smoking and lung cancer risk: a multicentric study from Europe. J Natl Cancer Inst 1999;91:697-701. [Crossref] [PubMed]
- Schiller JH, Harrington D, Belani C, et al. Comparison of four chemotherapy regimens for advanced non-small cell lung cancer. N Engl J Med 2002;346:92-8. [Crossref] [PubMed]
- Masters GA, Temin S, Azzoli CG, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2015;33:3488-515. [Crossref] [PubMed]
- Herbst RS, Johnson DH, Mininberg E, et al. Phase I/II trial evaluating the anti-vascular endothelial growth factor monoclonal antibody Bevacizumab in combination with the HER-1/Epidermal growth factor receptor tyrosine kinase inhibitor Erlotinib for patients with recurrent non-small cell lung cancer. J Clin Oncol 2005;23:2544-55. [Crossref] [PubMed]
- Johnson DH, Fehrencbacher L, Novotny WF, et al. Randomized phase II trial comparing Bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small cell lung cancer. J Clin Oncol 2004;22:2184-91. [Crossref] [PubMed]
- Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 2008;26:3543-51. [Crossref] [PubMed]
- Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019;16:341-55. [Crossref] [PubMed]
- Aggarwal C, Thompson JC, Black TA, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol 2019;5:173-80. [Crossref] [PubMed]
- Hierro C, Matos I, Martin-Liberal J, et al. Agnostic-Histology Approval of New Drugs in Oncology: Are We Already There? Clin Cancer Res 2019;25:3210-9. [Crossref] [PubMed]
- Yan L, Zhang W. Precision medicine becomes reality-tumor type-agnostic therapy. Cancer Commun (Lond) 2018;38:6. [Crossref] [PubMed]
- Sicklick JK, Kato S, Okamura R, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med 2019;25:744-50. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Rossi G, Pelosi G, Barbareschi M, et al. Subtyping non-small cell lung cancer: relevant issues and operative recommendations for the best pathology practice. Int J Surg Pathol 2013;21:326-36. [Crossref] [PubMed]
- Rossi G, Tiseo M, Cavazza A, et al. Is immunohistochemistry always required to diagnose lung cancer? Adv Anat Pathol 2013;20:327-33. [Crossref] [PubMed]
- Yatabe Y, Dacic S, Borczuk AC, et al. Best Practices Recommendations for Diagnostic Immunohistochemistry in Lung Cancer. J Thorac Oncol 2019;14:377-407. [Crossref] [PubMed]
- Rekhtman N, Ang DC, Sima CS, et al. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol 2011;24:1348-59. [Crossref] [PubMed]
- Pelosi G, Fabbri A, Bianchi F, et al. D (delta)Np63 (p40) and thyroid transcription factor-1 (TTF1) immunoreactivity upon small biopsies or cell blocks for typing non-small cell lung cancer: a novel two-hit, sparing-material approach. J Thorac Oncol 2012;7:281-90. [Crossref] [PubMed]
- Bishop JA, Teruya-Feldstein J, Westra WH, et al. p40 (DNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol 2012;25:405-15. [Crossref] [PubMed]
- Matoso A, Singh K, Jacob R, et al. Comparison of thyroid transcription factor-1 expression by 2 monoclonal antibodies in pulmonary and nonpulmonary primary tumors. Appl Immunohistochem Mol Morphol 2010;18:142-9. [Crossref] [PubMed]
- Rossi G, Longo L, Barbieri F, et al. Large cell neuroendocrine carcinoma of the lung: chemotherapy regimen depends on how "large" your diagnostic criteria are. Eur Respir J 2017; [Crossref] [PubMed]
- Rekhtman N, Pietanza MC, Hellmann MD, et al. Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma-like and Non-Small Cell Carcinoma-like Subsets. Clin Cancer Res 2016;22:3618-29. [Crossref] [PubMed]
- Derks JL, Leblay N, Thunnissen E, et al. Molecular Subtypes of Pulmonary Large-cell Neuroendocrine Carcinoma Predict Chemotherapy Treatment Outcome. Clin Cancer Res 2018;24:33-42. [Crossref] [PubMed]
- Rossi G, Cavazza A, Marchioni A, et al. Role of chemotherapy and the receptor tyrosine kinases KIT, PDGFRalpha, PDGFRbeta, and Met in large-cell neuroendocrine carcinoma of the lung. J Clin Oncol 2005;23:8774-85. [Crossref] [PubMed]
- Karlsson A, Brunnström H, Lindquist KE, et al. Mutational and gene fusion analyses of primary large cell and large cell neuroendocrine lung cancer. Oncotarget 2015;6:22028-37. [Crossref] [PubMed]
- Sholl LM. Large-cell carcinoma of the lung: a diagnostic category redefined by immunohistochemistry and genomics. Curr Opin Pulm Med 2014;20:324-31. [Crossref] [PubMed]
- Pelosi G, Barbareschi M, Cavazza A, et al. Large cell carcinoma of the lung: a tumor in search of an author. A clinically oriented critical reappraisal. Lung Cancer 2015;87:226-31. [Crossref] [PubMed]
- Rekhtman N, Tafe LJ, Chaft JE, et al. Distinct profile of driver mutations and clinical features in immunomarker-defined subsets of pulmonary large-cell carcinoma. Mod Pathol 2013;26:511-22. [Crossref] [PubMed]
- Driver BR, Portier BP, Mody DR, et al. Next-Generation Sequencing of a Cohort of Pulmonary Large Cell Carcinomas Reclassified by World Health Organization 2015 Criteria. Arch Pathol Lab Med 2016;140:312-7. [Crossref] [PubMed]
- Pelosi G, Fabbri A, Papotti M, et al. Dissecting Pulmonary Large-Cell Carcinoma by Targeted Next Generation Sequencing of Several Cancer Genes Pushes Genotypic-Phenotypic Correlations to Emerge. J Thorac Oncol 2015;10:1560-9. [Crossref] [PubMed]
- Rossi G, Mengoli MC, Cavazza A, et al. Large cell carcinoma of the lung: clinically oriented classification integrating immunohistochemistry and molecular biology. Virchows Arch 2014;464:61-8. [Crossref] [PubMed]
- Baldovini C, Rossi G, Ciarrocchi A. Approaches to Tumor Classification in Pulmonary Sarcomatoid Carcinoma. Lung Cancer (Auckl) 2019;10:131-49. [Crossref] [PubMed]
- Rossi G, Cavazza A, Sturm N, et al. Pulmonary carcinomas with pleomorphic, sarcomatoid, or sarcomatous elements: a clinicopathologic and immunohistochemical study of 75 cases. Am J Surg Pathol 2003;27:311-24. [Crossref] [PubMed]
- Fallet V, Saffroy R, Girard N, et al. High-throughput somatic mutation profiling in pulmonary sarcomatoid carcinomas using the LungCarta™ Panel: exploring therapeutic targets. Ann Oncol 2015;26:1748-53. [Crossref] [PubMed]
- Manzotti G, Torricelli F, Benedetta D, et al. An Epithelial-to-Mesenchymal Transcriptional Switch Triggers Evolution of Pulmonary Sarcomatoid Carcinoma (PSC) and Identifies Dasatinib as New Therapeutic Option. Clin Cancer Res 2019;25:2348-60. [Crossref] [PubMed]
- Tong JH, Yeung SF, Chan AW, et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin Cancer Res 2016;22:3048-56. [Crossref] [PubMed]
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discov 2015;5:850-9. [Crossref] [PubMed]
- Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 2015;16:e165-72. [Crossref] [PubMed]
- Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017;35:3065-74. [Crossref] [PubMed]
- Minari R, Bordi P, Del Re M, et al. Primary resistance to osimertinib due to SCLC transformation: Issue of T790M determination on liquid re-biopsy. Lung Cancer 2018;115:21-7. [Crossref] [PubMed]
- Ferrer L, Giaj Levra M, Brevet M, et al. A Brief Report of Transformation From NSCLC to SCLC: Molecular and Therapeutic Characteristics. J Thorac Oncol 2019;14:130-4. [Crossref] [PubMed]
- Marcoux N, Gettinger SN, O'Kane G, et al. EGFR-Mutant Adenocarcinomas That Transform to Small-Cell Lung Cancer and Other Neuroendocrine Carcinomas: Clinical Outcomes. J Clin Oncol 2019;37:278-85. [Crossref] [PubMed]
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol 2019;14:1784-93. [Crossref] [PubMed]
- Hsieh MS, Lin MW, Lee YH. Lung adenocarcinoma with sarcomatoid transformation after tyrosine kinase treatment and chemotherapy. Lung Cancer 2019;137:76-84. [Crossref] [PubMed]
- Chung JH, Rho JK, Xu X, et al. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer 2011;73:176-82. [Crossref] [PubMed]
- Xu S, Liu X, Liu R, et al. Concurrent epidermal growth factor receptor T790M secondary mutation and epithelial-mesenchymal transition in a lung adenocarcinoma patient with EGFR-TKI drug resistance. Thorac Cancer 2017;8:693-7. [Crossref] [PubMed]
- Xia L, Shao YW, Xia Y. Nkx2-4 Mutation Confers Resistance to EGFR-Tyrosine Kinase Inhibitors in EGFR-Mutant Lung Sarcomatoid Carcinoma. J Thorac Oncol 2019;14:e125-6. [Crossref] [PubMed]
- Sesumi Y, Suda K, Mizuuchi H, et al. Effect of dasatinib on EMT-mediated-mechanism of resistance against EGFR inhibitors in lung cancer cells. Lung Cancer 2017;104:85-90. [Crossref] [PubMed]
- Park S, Shim JH, Lee B, et al. Paired genomic analysis of squamous cell carcinoma transformed from EGFR-mutated lung adenocarcinoma. Lung Cancer 2019;134:7-15. [Crossref] [PubMed]
- Roca E, Pozzari M, Vermi W, et al. Outcome of EGFR-mutated adenocarcinoma NSCLC patients with changed phenotype to squamous cell carcinoma after tyrosine kinase inhibitors: A pooled analysis with an additional case. Lung Cancer 2019;127:12-8. [Crossref] [PubMed]
- Lin MW, Su KY, Su TJ, et al. Clinicopathological and genomic comparisons between different histologic components in combined small cell lung cancer and non-small cell lung cancer. Lung Cancer 2018;125:282-90. [Crossref] [PubMed]
- Shao Y, Zhong DS. Histological transformation after acquired resistance to epidermal growth factor tyrosine kinase inhibitors. Int J Clin Oncol 2018;23:235-42. [Crossref] [PubMed]
- Baglivo S, Ludovini V, Sidoni A, et al. Large Cell Neuroendocrine Carcinoma Transformation and EGFR-T790M Mutation as Coexisting Mechanisms of Acquired Resistance to EGFR-TKIs in Lung Cancer. Mayo Clin Proc 2017;92:1304-11. [Crossref] [PubMed]
- Shee-Chai C, Liam CK, Mun KS. Small Cell Transformation and T790M Mutation as Coresistance Mechanisms for First-line Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinase Inhibitor (TKI) Therapy Failure. J Thorac Oncol 2017;12:e171-3. [Crossref] [PubMed]
- Tokaca N, Wotherspoon A, Nicholson AG, et al. Lack of response to nivolumab in a patient with EGFR-mutant non-small cell lung cancer adenocarcinoma sub-type transformed to small cell lung cancer. Lung Cancer 2017;111:65-8. [Crossref] [PubMed]
- Xu Y, Huang Z, Gong L, et al. A case of resistance to tyrosine kinase inhibitor therapy: small cell carcinoma transformation concomitant with plasma-genotyped T790M positivity. Anticancer Drugs 2017;28:1056-61. [Crossref] [PubMed]
- Ou SI, Lee TK, Young L, et al. Dual occurrence of ALK G1202R solvent front mutation and small cell lung cancer transformation as resistance mechanisms to second generation ALK inhibitors without prior exposure to crizotinib. Pitfall of solely relying on liquid re-biopsy? Lung Cancer 2017;106:110-4. [Crossref] [PubMed]
- Le T, Sailors J, Oliver DH, et al. Histologic transformation of EGFR mutant lung adenocarcinoma without exposure to EGFR inhibition. Lung Cancer 2017;105:14-6. [Crossref] [PubMed]
- Hou S, Zhou S, Qin Z, et al. Evidence, Mechanism, and Clinical Relevance of the Transdifferentiation from Lung Adenocarcinoma to Squamous Cell Carcinoma. Am J Pathol 2017;187:954-62. [Crossref] [PubMed]
- Li L, Wang H, Li C, et al. Transformation to small-cell carcinoma as an acquired resistance mechanism to AZD9291: A case report. Oncotarget 2017;8:18609-14. [PubMed]
- Longo L, Mengoli MC, Bertolini F, et al. Synchronous occurrence of squamous-cell carcinoma "transformation" and EGFR exon 20 S768I mutation as a novel mechanism of resistance in EGFR-mutated lung adenocarcinoma. Lung Cancer 2017;103:24-6. [Crossref] [PubMed]
- Caumont C, Veillon R, Gros A, et al. Neuroendocrine phenotype as an acquired resistance mechanism in ALK-rearranged lung adenocarcinoma. Lung Cancer 2016;92:15-8. [Crossref] [PubMed]
- Miyamoto S, Ikushima S, Ono R, et al. Transformation to small-cell lung cancer as a mechanism of acquired resistance to crizotinib and alectinib. Jpn J Clin Oncol 2016;46:170-3. [PubMed]
- Abdallah N, Nagasaka M, Abdulfatah E, et al. Non-small cell to small cell lung cancer on PD-1 inhibitors: two cases on potential histologic transformation. Lung Cancer (Auckl) 2018;9:85-90. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26 [Crossref] [PubMed]
- Kato H, Ichinose Y, Ohta M, et al. A randomized trial of adjuvant chemotherapy with uracil-tegafur for adenocarcinoma of the lung. N Engl J Med 2004;350:1713-21. [Crossref] [PubMed]
- Georgoulias V, Papadakis E, Alexopoulos A, et al. Platinum-based and non-platinum-based chemotherapy in advanced non-small-cell lung cancer: a randomised multicentre trial. Lancet 2001;357:1478-84. [Crossref] [PubMed]
- Travis WD, Linnoila RI, Tsokos MG, et al. Neuroendocrine tumors of the lung with proposed criteria for large cell neuroendocrine carcinoma. An ultrastructural, immunohistochemical and flow cytometry study of 35 cases. Am J Surg Pathol 1991;15:529-53. [Crossref] [PubMed]
- Battafarano RJ, Fernandez FG, Ritter J, et al. Large cell neuroendocrine carcinoma: an aggressive form of non-small cell lung cancer. J Thorac Cardiovasc Surg 2005;130:166-72. [Crossref] [PubMed]
- Iyoda A, Hiroshima K, Toyozaki T, et al. Adjuvant chemotherapy for large cell carcinoma with neuroendocrine features. Cancer 2001;92:1108-12. [Crossref] [PubMed]
- Iyoda A, Hiroshima K, Moriya Y, et al. Prospective study of adjuvant chemotherapy for pulmonary large cell neuroendocrine carcinoma. Ann Thorac Surg 2006;82:1802-7. [Crossref] [PubMed]
- Grønberg BH, Bremnes RM, Fløtten O, et al. Phase III study by the Norwegian lung cancer study group: pemetrexed plus carboplatin compared with gemcitabine plus carboplatin as first-line chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol 2009;27:3217-24. [Crossref] [PubMed]
- Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med 2006;355:2542-50. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-237. Erratum in Ann Oncol 2019;30:863-70. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Riely GJ, Marks J, Pao W. KRAS Mutations in Non-Small Cell Lung Cancer. Proc Am Thorac Soc 2009;6:201-5. [Crossref] [PubMed]
- Pao W, Iafrate JA, Su Z. Genetically informed lung cancer medicine. J Pathol 2011;223:230-40. [Crossref] [PubMed]
- Yousem SA. Role of molecular studies in the diagnosis of lung adenocarcinoma. Mod Pathol 2012;25:S11-7. [Crossref] [PubMed]
- Tatematsu A, Shimizu J, Murakami Y, et al. Epidermal growth factor receptor mutations in small cell lung cancer. Clin Cancer Res 2008;14:6092-6. [Crossref] [PubMed]
- Fukui T, Tsuta K, Furuta K, et al. Epidermal growth factor receptor mutation status and clinicopathological features of combined small cell carcinoma with adenocarcinoma of the lung. Cancer Sci 2007;98:1714-9. [Crossref] [PubMed]
- Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006;355:213-5. [Crossref] [PubMed]
- Leone A, Graziano P, Gasbarra R, et al. Identification of EGFR mutations in lung sarcomatoid carcinoma. Int J Cancer 2011;128:732-5. [Crossref] [PubMed]
- Shukuya T, Takahashi T, Kaira R, et al. Efficacy of gefitinib for non-adenocarcinoma non-small-cell lung cancer patients harbouring epidermal growth factor receptor mutations: a pooled analysis of published reports. Cancer Sci 2011;102:1032-7. [Crossref] [PubMed]
- The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine. (NGM). A genomics-based classification of human lung tumors. Sci Transl Med 2013;5:209ra153 [PubMed]
- Rekhtman N. Commentary on Testing of Non-Adenocarcinomas. Arch Pathol Lab Med 2018;142:798. [Crossref] [PubMed]
- Langer CJ, Besse B, Gualberto A, et al. The evolving role of histology in the management of advanced non-small-cell lung cancer. J Clin Oncol 2010;28:5311-20. [Crossref] [PubMed]
- Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 Mutations. Clin Cancer Res 2012;18:1167-76. [Crossref] [PubMed]
- Lebanony D, Benjamin H, Gilad S, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol 2009;27:2030-7. [Crossref] [PubMed]
- Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 2001;98:13790-5. [Crossref] [PubMed]
- Wilkerson MD, Yin X, Hoadley KA, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res 2010;16:4864-75. [Crossref] [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with EGFR gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with nonsmall-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ou SHI, et al. ROS1 Rearrangements Define a Unique Molecular Class of Lung Cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Inamura K, Takeuchi K, Togashi Y, et al. EML4- ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 2008;3:13-7. [Crossref] [PubMed]
- Buttitta F, Barassi F, Fresu G, et al. Mutational analysis of the HER2 gene in lung tumors from Caucasian patients: mutations are mainly present in adenocarcinomas with bronchioloalveolar features. Int J Cancer 2006;119:2586-91. [Crossref] [PubMed]
- Rimkunas VM, Crosby KE, Kelly ME, et al. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res 2012;18:4449-57. [Crossref] [PubMed]
- Meyerson M, Franklin WA, Kelley MJ. Molecular classification and molecular genetics of human lung cancers. Semin Oncol 2004;31:4-19. [Crossref] [PubMed]
- Hagemann IS, O’Neill PK, Erill I, et al. Diagnostic yield of targeted next generation sequencing in various cancer types: an information-theoretic approach. Cancer Genet 2015;208:441-7. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014;158:929-44. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Chang JC, Alex D, Bott M, et al. Comprehensive Next-Generation Sequencing Unambiguously Distinguishes Separate Primary Lung Carcinomas From Intrapulmonary Metastases: Comparison with Standard Histopathologic Approach. Clin Cancer Res 2019;25:7113-25. [Crossref] [PubMed]
- Bennett CW, Berchem G, Kim YJ, et al. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget 2016;7:71013-35. [Crossref] [PubMed]
- Zhang YC, Zhou Q, Wu YL. The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. J Hematol Oncol 2017;10:167. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 2018;13:323-58. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Cai W, Lin D, Wu C, et al. Intratumoral heterogeneity of ALK-rearranged and ALK/EGFR coaltered lung adenocarcinoma. J Clin Oncol 2015;33:3701-9. [Crossref] [PubMed]
- Rijavec E, Coco S, Genova C, et al. Liquid Biopsy in Non-Small Cell Lung Cancer: Highlights and Challenges. Cancers (Basel) 2019; [Crossref] [PubMed]
- Rossi G, Bargellini I, Bonifazi M, et al. Optimized tumor sampling and processing by a multidisciplinary approach for an accurate diagnosis in non-small cell lung cancer. EMJ Oncol 2019;7:90-9.
- Chouaid C, Dujon C, Do P, et al. Feasibility and clinical impact of re-biopsy in advanced non-small-cell lung cancer: a prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer 2014;86:170-3. [Crossref] [PubMed]
- Heerink WJ, de Bock GH, de Jonge GJ, et al. Complication rates of CT-guided transthoracic lung biopsy: meta-analysis. Eur Radiol 2017;27:138-48. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Zhang B, Niu X, Zhang Q, et al. Circulating tumor DNA detection is correlated to histologic types in patients with early-stage non-small-cell lung cancer. Lung Cancer 2019;134:108-16. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Müller JN, Falk M, Talwar J, et al. Concordance between comprehensive cancer genome profiling in plasma and tumor specimens. J Thorac Oncol 2017;12:1503-11. [Crossref] [PubMed]
- Zhou M, Guan Y, Yang X, et al. The prognostic and therapeutic role of genomic subtyping by sequencing tumor or cell-free DNA in pulmonary large-cell neuroendocrine carcinoma. Clin Cancer Res 2020; [Crossref] [PubMed]
- Blumenthal GM, Pazdur R. Approvals in 2018: a histology-agnostic new molecular entity, novel end points and real-time review. Nat Rev Clin Oncol 2019;16:139-41. [Crossref] [PubMed]
- Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplex genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011;22:2616-24. [Crossref] [PubMed]
Cite this article as: Galli G, Rossi G. Lung cancer histology-driven strategic therapeutic approaches. Shanghai Chest 2020;4:29.


