
Core needle biopsy for screening detected lung cancer—does it capture all in light of tumor heterogeneity?—a narrative review
Introduction
Lung cancer is one of the most frequent cancers worldwide. It accounts for the majority of cancer deaths in men and is the second most frequent cause of cancer death in women (1).Several lung cancer entities are recognized by the WHO classification with adenocarcinoma, squamous cell carcinoma and small cell carcinoma representing the most prevalent entities (2). The histologic differentiation of lung carcinomas is associated with prognoses and therapeutic consequences (3,4). Areas with varying differentiation that correspond to different entities, e.g., with squamous and adenocarcinoma differentiation, may be found within a lung cancer (5). Additionally, amongst adenocarcinomas morphological variants are recognized that can be present in different areas of a single tumor. Lepidic, acinar, papillary, micropapillary and solid patterns are seen most frequently (2). TheWHO classification of lung tumors recommends to report the different morphological subtypes present in lung adenocarcinoma resection specimens and to specify the predominant histologic subtype (2), which correlates with overall and disease free survival and may predict the benefit from adjuvant chemo- and radiotherapy (6,7).
Adenocarcinoma, squamous cell carcinoma and small cell carcinoma are genetically different diseases (8,9). Additionally, within a given lung cancer genetic intratumoral heterogeneity is prevalent (10,11). Moreover, the genetic composition of a primary lung carcinoma and its metastasis may differ (12). Tumor heterogeneity, however, is not restricted to the genetic makeup of cancer cells but rather involves also differences in protein expression as well. These differences may be influenced by intrinsic and extrinsic factors, such as interferon-γ release by immune cells in the case of PD-L1, a molecular target for immune checkpoint inhibitor therapy in lung cancer (13).
A lung nodule implies a pulmonary lesion that is well-circumscribed and 3 cm or less in diameter. In contrast, the terms tumor or mass are used for lesions larger than 3 cm (14). In case imaging detected a lung nodule or tumor in symptomatic patients or accidentally revealed one in asymptomatic patients or in the course of a screening program, in particular with low dose chest computed tomography (CT), further clarification of the lesion is required (15-17). The options include surveillance CT imaging, CT-guided biopsy, transbronchial biopsy for central lesions, and surgical resection (18). However, most lung cancers are in an advanced and inoperable stage at diagnosis. Therefore, diagnosis relies heavily on obtaining a sufficient number of tumor cells for cytology and/or on retrieving small pieces of tumor tissue for morphological diagnosis and molecular testing to identify drug targets. To this end, tissue from a peripheral nodule or tumor is frequently retrieved by image guided (mostly by using CT) core needle biopsy (CNB) (18-20). With tumor heterogeneity in mind, the question arises whether such a biopsy is representative for the tumor and whether it leads to the correct diagnosis and molecular test results which form the basis for the selection of the most appropriate therapy. In this review, we will summarize the strengths and limitations of core needle biopsies for the diagnosis of lung cancer, histological subtyping and molecular testing.
We present the following article in accordance with the Narrative Review reporting checklist (available at: https://dx.doi.org/10.21037/shc-21-1).
Methods
We searched the literature using PubMed for articles on CNB, lung cancer classification, morphologic subtypes of lung cancer, lung cancer biomarkers, tumor heterogeneity and genetics of lung cancer. Relevant publications were reviewed by the authors and the most pertinent results summarized.
CNB and morphologic heterogeneity of lung cancer
A lung nodule or tumor detected during screening can be further evaluated by transthoracic fine-needle aspiration biopsy (FNAB) or CNB. Both procedures are more commonly guided by CT and less frequently by ultrasound, the latter being employed for peripheral lesions abutting the pleura (19,21,22). Pooled data from 48 studies demonstrate a high diagnostic accuracy for both methods. The overall diagnostic accuracy of CT-guided biopsy (FNAB plus CNB) was 92.1% with a sensitivity of 92.1% and a specificity of around 100% for the diagnosis of malignancy (23). CNB has a slightly higher sensitivity, specificity and accuracy for detecting a malignancy in comparison with FNAB (21,22,24,25). Even for small nodules with a diameter ≤2 cm the diagnostic accuracy of CNB ranged from 87–95% (18). Whereas FNAB provides only cytology specimens for smears and/or cell blocks, CNB yields a tissue specimen. Although FNAB is a useful technique for the diagnosis of lung cancer, CNB provides the advantage of facilitating a histology with preservation of the architectural context of lesional cells and their microenvironment. FNAB and CNB are regarded as rather safe procedures, even though complications may occur, with pneumothorax and pulmonary hemorrhage being the most frequent and found at rates of 20.5% and 2.8%, respectively, in a pooled analysis of 75 publications (23).
Adenocarcinomas are heterogeneous in their morphology with frequently different growth patterns present within a single tumor (Figure 1). In contrast, squamous cell carcinoma and neuroendocrine carcinoma show less morphologic variation within a given tumor, although mixed tumors, e.g., adenosquamous carcinomas or mixed neuroendocrine/non-neuroendocrine carcinomas and biphasic epithelial-mesenchymal tumors (carcinosarcomas) do occur (26). The WHO classification recommends for lung adenocarcinomas a semiquantitative estimation of each growth pattern in 5% increments for determining the single predominant differentiation (2). The pattern of differentiation has prognostic impact. For example, the presence of a micropapillary component of 5% or greater is a risk factor for recurrence in patients treated with limited resection (27). Furthermore, in patients with stage I lung adenocarcinoma, a solid predominant subtype is an independent predictor of early recurrence (28).The morphologic pattern may also provide predictive information on response to therapy. For instance, in early stage lung adenocarcinomas the response to stereotactic body radiation therapy was better in tumors with lepidic, acinar and papillary differentiation than in micropapillary and/or solid pattern carcinomas (7). Moreover, patients with micropapillary and solid patterns benefit more from adjuvant chemotherapy (4). In spite of these observations, therapy stratification according to the predominant histologic subtype is currently not a standardized approach, yet, it may be important for prospective clinical trial designs.
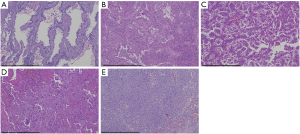
In case of CNB the question arises whether the biopsy is predictive of the predominant morphologic subtype of the whole lung nodule or tumor. Matsuzawa et al. reported a 66% concordance of predominant subtypes in 326 biopsies and matched surgical resection specimens (29). As might be expected, histopathologic diagnostic concordance and the extent of tumor area captured by the biopsy correlated. Biopsies with tumor areas >0.7 mm2 were slightly higher concordant than biopsies with smaller tumor areas (<0.7 mm2) (29). A similar concordance rate between CNB and surgically resected tumors of overall 64% was observed by Tsai et al., however, the concordance differed among subtypes with lepidic 78% (91/117); acinar 58% (98/168); papillary 33% (1/3); micropapillary 80% (4/5); and solid 44% (11/25) concordance (25). There was no difference in the concordance rate associated with the punctured part of the tumor (central or peripheral areas within the tumor) and the number of times a puncture was performed. In line with the reports of Matsuzawa and Tsai the study of Huang et al. observed in 58.6% of their 128 cases concordant predominant pattern subtyping of adenocarcinoma between biopsy and the corresponding surgical resection specimen (30).
Although lobectomy is the therapy of choice for early stage lung carcinomas, in selected cases, e.g., patients with a contraindication for lobar resection or node-negative adenocarcinomas with a low-grade growth pattern, a limited resection would be advantageous as pulmonary tissue would be preserved (31,32). However, Huang et al. reported that only 18–33% morphologically high-grade (micropapillary/solid predominant) tumors were detected by CNB (30). Therefore, CNB cannot predict reliably the presence or absence of high-grade adenocarcinoma predominance in a lung nodule or tumor and thus is not suitable to support therapeutic decisions depending on an exact subtype classification.
The ability to correctly determine the subtype of lung carcinoma and to evaluate the presence of invasion is lower with CNB tissues than with surgical resection specimens (33). The WHO classification recommends to diagnose adenocarcinoma and squamous cell carcinoma on small biopsies when the criteria for specific diagnosis of these entities are fulfilled (34). If clear adeno- or squamous cell patterns are absent, but immunohistochemistry (e.g., TTF-1 or p63 staining) favors either of them, the nomenclature “non-small cell carcinoma (NSCC), favor adenocarcinoma” or “NSCC, favor squamous cell carcinoma” should be used (34). A similar cautious naming has been suggested for CNB with histologies suggestive of large cell neuroendocrine carcinoma, adenosquamous carcinoma and sarcomatoid carcinoma (2,33).
A further limitation of CNB is the impossibility to definitely render the diagnosis of adenocarcinoma in-situ (AIS) or minimal invasive adenocarcinoma (MIA), as due to tumor heterogeneity an invasion may not be present in the biopsy. Therefore, a non-invasive growth pattern in a CNB should be referred to as a lepidic growth pattern (33,34). In the corresponding surgical resection specimen, the tumor may represent AIS, MIA or invasive adenocarcinoma with a lepidic component. Additionally, large cell carcinoma, an undifferentiated non-small-cell lung cancer (NSCLC), cannot be diagnosed by CNB. In a CNB specimen it should be named NSCLC-not otherwise specified (NOS). A definite diagnosis of large cell carcinoma requires a thoroughly sampled surgically resected tumor (33).
CNB and diagnostic accuracy of PD-L1 immunohistology
Immunotherapy with antibodies to PD-1 (programmed cell death protein 1) and its ligand PD-L1 is an option for the treatment of advanced NSCLC (35). The expression of PD-L1 on tumor cells is a biomarker that is enriched in patients that are more likely to respond to immunotherapy (36,37). The determination of the so-called tumor proportion score (TPS), which is the percentage of PD-L1 reactive cancer cells as assessed by immunohistochemistry, is essential for patient stratification (38). Patients with NSCLC with a TPS ≥50% are eligible for first-line treatment with the PD-1 blocking antibody pembrolizumab (35). In later therapy lines, depending on the therapeutic antibody used, the TPS cut-off level for immune therapy eligibility is only 1% or the proof of tumor PD-L1 expression is not mandatory at all, in particular in combination with chemotherapy (39,40).
The expression of PD-L1 by cancer cells is often spatially heterogenous within a tumor and may differ between the primary tumor and metastasis (41-43). Additionally, a temporal heterogeneity of PD-L1 expression exists, which is caused by tumor cell intrinsic mechanisms and external stimuli, such as interferon-γ released by immune cells (13). This spatial and temporal heterogeneity limits the validity of PD-L1 expression as a biomarker for immunotherapy.
The small amount of tumor tissue retrievable by CNB may confine the representativity of PD-L1 assessment. Tsai et al. reported that CNB specimens were adequate for immunohistochemical evaluation of PD-L1 expression in 96.4% (44). They retrieved a mean number of 7.9 biopsies per sample, which is above the more widely used 2–4 passes performed at most institutions (44). A limitation of the study of Tsai et al. is the lack of comparison of CNB PD-L1 stainings with corresponding surgically resected nodules or tumors. Gradecki et al. compared PD-L1 expression in 51 paired CNB and surgical resection specimens of NSCLC (45). PD-L1 staining was grouped as 0%, 1% to 49%, and ≥50% of carcinoma cells positive. In the PD-L1 strongly expressing group (≥50%) the concordance was high with 92.2%. The authors thus suggested that CNB may be adequate for determining PD-L1 in NSCLC. However, overall the CNB and the corresponding surgical resection specimen results were concordant in only 76% of cases, with discrepancies mainly in the low expressing cancers (45). Discordant PD-L1 scoring between biopsies and surgical resection specimens were also noted in other studies with false negative PD-L1 expression assessment with biopsies in 2.5% to 48% of cases (46-48). A low-level expression of PD-L1 may thus be missed with CNB, and these patients may be unwarrantedly excluded from immunotherapy. Therefore, it seems advisable to repeat the PD-L1 staining provided a surgical resection specimen is available, in particular for low-level expressing CNB specimens.
CNB and molecular heterogeneity of lung cancer
Lung adenocarcinoma, squamous cell carcinoma and small cell carcinoma are genetically different entities (8,9). Furthermore, each carcinoma exhibits a spectrum of mutated genes that differs between individual patients with a tumor of the same histologic subtype (49). Additionally, within a given tumor different areas harbor common as well as distinct mutations (10,11). This intratumor heterogeneity is the result of genetic instability of cancer cells and a selection of the fittest clones by evolution (50).Intratumor heterogeneity is in general regarded as a cause for the development of therapy resistance and may be associated with increased mortality (51,52). Regional exome sequencing of NSCLC revealed a branched model of cancer development with a trunk of mutations that are ubiquitous in a tumor and branches of mutations present only in subclones in some areas of a tumor (10,11). In the study of de Bruin et al. 13 of 14 known cancer gene mutations were present in the trunks of the mutation trees in 11 analyzed NSCLC (10). This indicates that these clonal mutations occurred early during evolution of the tumors. However, about a third of all nonsilent mutations were present in at least one region but not in all regions of individual tumors. Similar observations were made by Zhang et al. who reported 20 out of 21 known cancer driver gene mutations present in all regions of individual NSCLCs (11). Additionally, they observed that not all driver mutations develop before subclonal diversification. Some may arise thereafter. On average, 24% of all mutations were not present in all analyzed regions of a tumor and thus subclonal. A molecular heterogeneity is already present in the early in-situ and minimal invasive forms of lung adenocarcinomas (AIS and MIA), although key driver genes such as EGFR and KRAS are usually present in all regions of a tumor (53). In conclusion, although NSCLC exhibits high intratumoral genetic heterogeneity, single-area sequencing may be adequate to identify the majority of known key driver gene mutations in NSCLC at the time of initial tumor diagnosis (10,11).
A few studies addressed the question whether within a lung adenocarcinoma areas with different histologic differentiation harbor the same mutations. Dietz et al. investigated by digital PCR the distribution of allele frequencies of EGFR and KRAS mutations in lung adenocarcinomas in correlation to the morphological patterns in a tumor (54). Mutant allele frequencies were higher in areas with a predominant solid pattern compared to all other histologies. Nevertheless, their data indicate that driver gene mutations are present throughout the whole tumor, with a correlation between the allele frequencies and histologic growth patterns (54), which may be caused by differences in tumor cell abundancy. Mattsson et al. also reported consistent EGFR and KRAS mutation status within histologically heterogeneous lung cancers (55). Conversely, rare discrepancies in EGFR and KRAS mutations between areas with different growth patterns were observed by Zhong et al., who utilized direct sequencing and a mutation specific polymerase chain reaction (PCR) assay (56). They concluded that discrepancies might be due to the various abundance of mutant tumor cells and the sensitivity of the employed detection assay. In contrast, Tomonaga et al. observed a high intratumor EGFR mutation heterogeneity of 23.7% (9/38) between morphologically different areas in lung adenocarcinomas by employing a mutant-enriched PCR assay (57).
When considering possible differences in the mutation status between primary lung cancer and their metastasis contradictory results exist. For example, the percentage of discordant results of EGFR mutation status varied from 0% to 38.8% in 7 studies (58). A meta-analysis of 9 publications reported a 12.2% (86/707) discrepancy in EGFR mutation status between the primary lung carcinoma and lymph node metastasis (59). In contrast, other authors showed that mutation heterogeneity is rare between primary tumors and their metastasis. Kim et al. identified infrequent genetic heterogeneity of 16 genes, including EGFR, between primary tumor and metastasis by next-generation sequencing (12). Likewise, Vignot et al. also observed a high concordance of NGS detected recurrent somatic alterations between primary tumor and metastasis from patients with NSCLC (60). Sherwood et al. described in a review of 26 articles that there are variable discordance rates between primary tumor and their metastasis. However, as there is a substantial concordance, the authors concluded that molecular diagnosis could be made with the primary tumor or the metastasis, provided sensitive detection methods are used (61).
Genetic intratumor heterogeneity raises the question whether a small amount of tumor tissue retrieved by CNB is adequate for molecular diagnostics. The number of cells required for a successful mutation test has not been well defined, but a range of 100–400 tumor cells in the specimen has been suggested (58). Core needle biopsies provided an adequate amount of tissue to perform molecular studies in 100% of cases in one study compared to 94% of specimen retrieved by FNA (21). Solomon et al. observed 100% concordance in EGFR and KRAS testing in 16 lung adenocarcinomas between CNB and the surgical resection specimens (62). Likewise, Kim et al. compared EGFR, KRAS, BRAF and PIK3CA mutational status between 14 CNB and surgical resection specimens and did not observe any differences (63). Furthermore, Masago et al. compared EGFR exon 18, 19, 21 sequencing of biopsies with surgical resection specimens and found a 100% concordance in 18 paired samples (64). These data suggest that CNB obtained tissues may be sufficient to identify key driver gene mutations in lung adenocarcinomas. In line with that, according to the College of American Pathologists (CAP)/International Association for the Study of Lung Cancer (IASLC)/Association for Molecular Pathology (AMP) guideline on molecular testing in NSCLC, EGFR testing of multiple different areas within a single tumor is not necessary (65).
Conclusions
CNB is a highly valuable technique for the clarification of lung nodules and tumors with a high accuracy for the diagnosis of malignancy. Additionally, it can provide sufficient and suitable tissue for molecular testing. Limitations are posed by intratumor heterogeneity for the morphological subtyping of lung adenocarcinomas and PD-L1 assessment, in particular for low-level expressing tumors.
Acknowledgments
We are very grateful to Ms. Gertrude Krainz for English proofreading.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Wieland Voigt and Helmut Prosch) for the series “Lung Cancer Screening” published in Shanghai Chest. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at: https://dx.doi.org/10.21037/shc-21-1
Peer Review File: Available at https://dx.doi.org/10.21037/shc-21-1
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/shc-21-1). The series “Lung Cancer Screening” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict provision that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke AP, et al. editors. WHO classification of tumours of the lung, pleura, thymus and heart. IARC Press, Lyon; 2015.
- Russell PA, Wainer Z, Wright GM, et al. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 2011;6:1496-504. [Crossref] [PubMed]
- Russell PA, Wright GM. Predominant histologic subtype in lung adenocarcinoma predicts benefit from adjuvant chemotherapy in completely resected patients: discovery of a holy grail? Ann Transl Med 2016;4:16. [PubMed]
- de Sousa VML, Carvalho L. Heterogeneity in Lung Cancer. Pathobiology 2018;85:96-107. [Crossref] [PubMed]
- Hung JJ, Yeh YC, Jeng WJ, et al. Predictive value of the international association for the study of lung cancer/American Thoracic Society/European Respiratory Society classification of lung adenocarcinoma in tumor recurrence and patient survival. J Clin Oncol 2014;32:2357-64. [Crossref] [PubMed]
- Leeman JE, Rimner A, Montecalvo J, et al. Histologic Subtype in Core Lung Biopsies of Early-Stage Lung Adenocarcinoma is a Prognostic Factor for Treatment Response and Failure Patterns After Stereotactic Body Radiation Therapy. Int J Radiat Oncol Biol Phys 2017;97:138-45. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. [Crossref] [PubMed]
- Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256-9. [Crossref] [PubMed]
- Kim EY, Cho EN, Park HS, et al. Genetic heterogeneity of actionable genes between primary and metastatic tumor in lung adenocarcinoma. BMC Cancer 2016;16:27. [Crossref] [PubMed]
- Lamberti G, Sisi M, Andrini E, et al. The Mechanisms of PD-L1 Regulation in Non-Small-Cell Lung Cancer (NSCLC): Which Are the Involved Players? Cancers (Basel) 2020;12:3129. [Crossref] [PubMed]
- Hansell DM, Bankier AA, MacMahon H, et al. Fleischner Society: glossary of terms for thoracic imaging. Radiology 2008;246:697-722. [Crossref] [PubMed]
- Lopes Pegna A, Picozzi G, Falaschi F, et al. Four-year results of low-dose CT screening and nodule management in the ITALUNG trial. J Thorac Oncol 2013;8:866-75. [Crossref] [PubMed]
- de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N Engl J Med 2020;382:503-13. [Crossref] [PubMed]
- Lam S, Bryant H, Donahoe L, et al. Management of screen-detected lung nodules: A Canadian partnership against cancer guidance document. Canadian Journal of Respiratory, Critical Care, and Sleep Medicine 2020;4:236-65. [Crossref]
- Andrade JR, Rocha RD, Falsarella PM, et al. CT-guided percutaneous core needle biopsy of pulmonary nodules smaller than 2 cm: technical aspects and factors influencing accuracy. J Bras Pneumol 2018;44:307-14. [Crossref] [PubMed]
- Yao X, Gomes MM, Tsao MS, et al. Fine-needle aspiration biopsy versus core-needle biopsy in diagnosing lung cancer: a systematic review. Curr Oncol 2012;19:e16-27. [Crossref] [PubMed]
- Wang Y, Li W, He X, et al. Computed tomography-guided core needle biopsy of lung lesions: Diagnostic yield and correlation between factors and complications. Oncol Lett 2014;7:288-94. [Crossref] [PubMed]
- Coley SM, Crapanzano JP, Saqi A. FNA, core biopsy, or both for the diagnosis of lung carcinoma: Obtaining sufficient tissue for a specific diagnosis and molecular testing. Cancer Cytopathol 2015;123:318-26. [Crossref] [PubMed]
- Sangha BS, Hague CJ, Jessup J, et al. Transthoracic Computed Tomography-Guided Lung Nodule Biopsy: Comparison of Core Needle and Fine Needle Aspiration Techniques. Can Assoc Radiol J 2016;67:284-9. [Crossref] [PubMed]
- DiBardino DM, Yarmus LB, Semaan RW. Transthoracic needle biopsy of the lung. J Thorac Dis 2015;7:S304-16. [PubMed]
- Anzidei M, Porfiri A, Andrani F, et al. Imaging-guided chest biopsies: techniques and clinical results. Insights Imaging 2017;8:419-28. [Crossref] [PubMed]
- Tsai PC, Yeh YC, Hsu PK, et al. CT-Guided Core Biopsy for Peripheral Sub-solid Pulmonary Nodules to Predict Predominant Histological and Aggressive Subtypes of Lung Adenocarcinoma. Ann Surg Oncol 2020;27:4405-12. [Crossref] [PubMed]
- Ruffini E, Rena O, Oliaro A, et al. Lung tumors with mixed histologic pattern. Clinico-pathologic characteristics and prognostic significance. Eur J Cardiothorac Surg 2002;22:701-7. [Crossref] [PubMed]
- Nitadori J, Bograd AJ, Kadota K, et al. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or smaller. J Natl Cancer Inst 2013;105:1212-20. [Crossref] [PubMed]
- Ujiie H, Kadota K, Chaft JE, et al. Solid Predominant Histologic Subtype in Resected Stage I Lung Adenocarcinoma Is an Independent Predictor of Early, Extrathoracic, Multisite Recurrence and of Poor Postrecurrence Survival. J Clin Oncol 2015;33:2877-84. [Crossref] [PubMed]
- Matsuzawa R, Kirita K, Kuwata T, et al. Factors influencing the concordance of histological subtype diagnosis from biopsy and resected specimens of lung adenocarcinoma. Lung Cancer 2016;94:1-6. [Crossref] [PubMed]
- Huang KY, Ko PZ, Yao CW, et al. Inaccuracy of lung adenocarcinoma subtyping using preoperative biopsy specimens. J Thorac Cardiovasc Surg 2017;154:332-339.e1. [Crossref] [PubMed]
- Landreneau RJ, Normolle DP, Christie NA, et al. Recurrence and survival outcomes after anatomic segmentectomy versus lobectomy for clinical stage I non-small-cell lung cancer: a propensity-matched analysis. J Clin Oncol 2014;32:2449-55. [Crossref] [PubMed]
- Sesti J, Donington JS. Sublobar Resection: Ongoing Controversy for Treatment for Stage I Non-Small Cell Lung Cancer. Thorac Surg Clin 2016;26:251-9. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. Diagnosis of lung cancer in small biopsies and cytology: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch Pathol Lab Med 2013;137:668-84. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133-50. [Crossref] [PubMed]
- Pharaon R, Koczywas MA, Salgia S, et al. Biomarkers in immunotherapy: literature review and future directions. J Thorac Dis 2020;12:5119-27. [Crossref] [PubMed]
- Hutarew G. PD-L1 testing, fit for routine evaluation? From a pathologist's point of view. Memo 2016;9:201-6. [Crossref] [PubMed]
- Wang C, Kulkarni P, Salgia R. Combined Checkpoint Inhibition and Chemotherapy: New Era of 1st-Line Treatment for Non-Small-Cell Lung Cancer. Mol Ther Oncolytics 2019;13:1-6. [Crossref] [PubMed]
- Geraci E, Chablani L. Immunotherapy as a second-line or later treatment modality for advanced non-small cell lung cancer: A review of safety and efficacy. Crit Rev Oncol Hematol 2020;152:103009 [Crossref] [PubMed]
- Mansfield AS, Aubry MC, Moser JC, et al. Temporal and spatial discordance of programmed cell death-ligand 1 expression and lymphocyte tumor infiltration between paired primary lesions and brain metastases in lung cancer. Ann Oncol 2016;27:1953-8. [Crossref] [PubMed]
- Pinato DJ, Shiner RJ, White SD, et al. Intra-tumoral heterogeneity in the expression of programmed-death (PD) ligands in isogeneic primary and metastatic lung cancer: Implications for immunotherapy. Oncoimmunology 2016;5:e1213934 [Crossref] [PubMed]
- Uruga H, Bozkurtlar E, Huynh TG, et al. Programmed Cell Death Ligand (PD-L1) Expression in Stage II and III Lung Adenocarcinomas and Nodal Metastases. J Thorac Oncol 2017;12:458-66. [Crossref] [PubMed]
- Tsai EB, Pomykala K, Ruchalski K, et al. Feasibility and Safety of Intrathoracic Biopsy and Repeat Biopsy for Evaluation of Programmed Cell Death Ligand-1 Expression for Immunotherapy in Non-Small Cell Lung Cancer. Radiology 2018;287:326-32. [Crossref] [PubMed]
- Gradecki SE, Grange JS, Stelow EB. Concordance of PD-L1 Expression Between Core Biopsy and Resection Specimens of Non-Small Cell Lung Cancer. Am J Surg Pathol 2018;42:1090-4. [Crossref] [PubMed]
- Kitazono S, Fujiwara Y, Tsuta K, et al. Reliability of Small Biopsy Samples Compared With Resected Specimens for the Determination of Programmed Death-Ligand 1 Expression in Non--Small-Cell Lung Cancer. Clin Lung Cancer 2015;16:385-90. [Crossref] [PubMed]
- Ilie M, Long-Mira E, Bence C, et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol 2016;27:147-53. [Crossref] [PubMed]
- Gniadek TJ, Li QK, Tully E, et al. Heterogeneous expression of PD-L1 in pulmonary squamous cell carcinoma and adenocarcinoma: implications for assessment by small biopsy. Mod Pathol 2017;30:530-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Turajlic S, Sottoriva A, Graham T, et al. Resolving genetic heterogeneity in cancer. Nat Rev Genet 2019;20:404-16. [Crossref] [PubMed]
- Andor N, Graham TA, Jansen M, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016;22:105-13. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Izumchenko E, Chang X, Brait M, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun 2015;6:8258. [Crossref] [PubMed]
- Dietz S, Harms A, Endris V, et al. Spatial distribution of EGFR and KRAS mutation frequencies correlates with histological growth patterns of lung adenocarcinomas. Int J Cancer 2017;141:1841-8. [Crossref] [PubMed]
- Mattsson JS, Imgenberg-Kreuz J, Edlund K, et al. Consistent mutation status within histologically heterogeneous lung cancer lesions. Histopathology 2012;61:744-8. [Crossref] [PubMed]
- Zhong WZ, Su J, Xu FP, et al. Rare discrepancies in a driver gene alteration within histologically heterogeneous primary lung cancers. Lung Cancer 2015;90:205-11. [Crossref] [PubMed]
- Tomonaga N, Nakamura Y, Yamaguchi H, et al. Analysis of intratumor heterogeneity of EGFR mutations in mixed type lung adenocarcinoma. Clin Lung Cancer 2013;14:521-6. [Crossref] [PubMed]
- Kim L, Tsao MS. Tumour tissue sampling for lung cancer management in the era of personalised therapy: what is good enough for molecular testing? Eur Respir J 2014;44:1011-22. [Crossref] [PubMed]
- Wang F, Fang P, Hou DY, et al. Comparison of epidermal growth factor receptor mutations between primary tumors and lymph nodes in non-small cell lung cancer: a review and meta-analysis of published data. Asian Pac J Cancer Prev 2014;15:4493-7. [Crossref] [PubMed]
- Vignot S, Frampton GM, Soria JC, et al. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol 2013;31:2167-72. [Crossref] [PubMed]
- Sherwood J, Dearden S, Ratcliffe M, et al. Mutation status concordance between primary lesions and metastatic sites of advanced non-small-cell lung cancer and the impact of mutation testing methodologies: a literature review. J Exp Clin Cancer Res 2015;34:92. [Crossref] [PubMed]
- Solomon SB, Zakowski MF, Pao W, et al. Core needle lung biopsy specimens: adequacy for EGFR and KRAS mutational analysis. AJR Am J Roentgenol 2010;194:266-9. [Crossref] [PubMed]
- Kim HK, Lee HY, Choi YL, et al. Assessment of intratumoral heterogeneity of oncogenic driver mutations in surgically-resected lung adenocarcinoma: implications of percutaneous biopsy-based molecular assay for target-directed therapy. Anticancer Res 2014;34:707-14. [PubMed]
- Masago K, Fujita S, Mio T, et al. Accuracy of epidermal growth factor receptor gene mutation analysis by direct sequencing method based on small biopsy specimens from patients with non-small cell lung cancer: analysis of results in 19 patients. Int J Clin Oncol 2008;13:442-6. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. [Crossref] [PubMed]
Cite this article as: Binder C, Oberndorfer F, Müllauer L. Core needle biopsy for screening detected lung cancer—does it capture all in light of tumor heterogeneity?—a narrative review. Shanghai Chest 2021;5:40.


